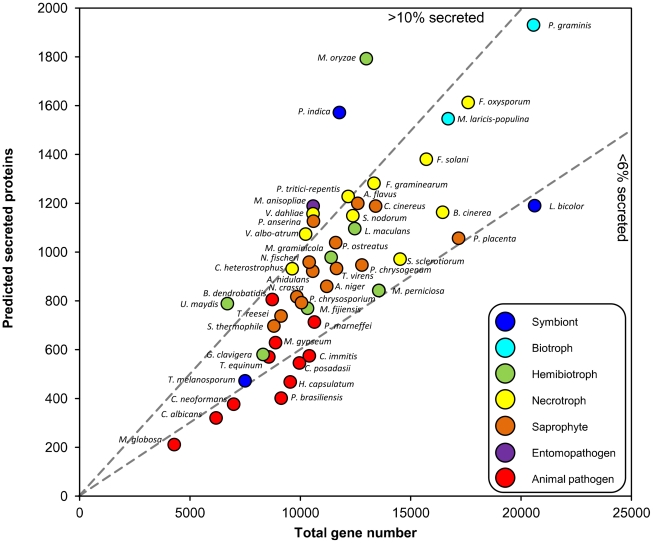
Figure 1. Relationship between predicted secreted protein number and total gene content of fungi.
Data are from [3], or by applying SignalP to genome releases (indicated by *). Dashed lines discriminate between fungi with high (>10) or low (<6) % secreted proteins. Animal pathogens: Batrachochytrium dendrobatidis, Candida albicans, Coccidioides immitis, C. posadasii, Cryptococcus neoformans, Histoplasma capsulatum, Malassezia globosa, Microsporum gypseum, Paracoccidioides brasiliensis, Penicillium marneffei, Trichophyton equinum. Hemibiotrophs: Grosmania clavigera*, Leptosphaeria maculans* [14], Magnaporthe oryzae*, Mycosphaerella fijiensis, M. graminicola, Moniliophthora perniciosa, Ustilago maydis. Entomopathogen: Metarhizium anisopliae* [4]. Necrotrophs: Botrytis cinerea, Cochliobolus heterostrophus, Fusarium graminearum, F. oxysporum, F. solani, Pyrenophora tritici-repentis, Stagonospora nodorum*, Sclerotinia sclerotiorum, Verticillium albo-atrum, V. dahliae. Biotrophs: Melampsora laricis-populina* [15], Puccinia graminis f.sp. tritici. Saprophytes: Aspergillus flavus, A. nidulans, A. niger, Coprinus cinereus, Neurospora crassa, Neosartorya fischeri, Podospora anserina, Penicillium chrysogenum, Phanerochaete chrysosporium, Pleurotus ostreatus, Postia placenta, Sporotrichum thermophile, Trichoderma reesei, T. virens. Symbionts: Laccaria bicolor, Piriformospora indica* [3], Tuber melanosporum* [16].