Abstract
MicroRNAs (miRNAs) are involved in post-transcriptional regulation of gene expression through binding to messenger RNAs (mRNA) thereby promoting mRNA degradation or altered translation. A single-nucleotide polymorphism (SNP) located within a miRNA-binding site could thus alter mRNA translation and influence cancer risk and treatment response. The common SNPs located within the 3′-untranslated regions of 20 DNA repair genes were analysed for putative miRNA-binding sites using bioinformatics algorithms, calculating the difference in Gibbs free binding energy (ΔΔG) for each wild-type versus variant allele. Seven SNPs were selected to be genotyped in germ line DNAs both from a bladder cancer case–control series (752 cases and 704 controls) and 202 muscle-invasive bladder cancer radiotherapy cases. The PARP-1 SNP rs8679 was also genotyped in a breast cancer case–control series (257 cases and 512 controls). Without adjustment for multiple testing, multivariate analysis demonstrated an association with increased bladder cancer risk with PARP1 rs8679 (Ptrend = 0.05) while variant homozygotes of PARP1 rs8679 were also noted to have an increased breast cancer risk (P = 0.03). In the radiotherapy cases, carriers of the RAD51 rs7180135 minor allele had improved cancer-specific survival (hazard ratio 0.52, 95% confidence interval 0.31–0.87, P = 0.01). This is the first report of associations between DNA repair gene miRNA-binding site SNPs with bladder and breast cancer risk and radiotherapy outcomes. If validated, these findings may give further insight into the biology of bladder carcinogenesis, allow testing of the RAD51 SNP as a potential predictive biomarker and also reveal potential targets for new cancer treatments.
Introduction
In recent years, there has been increasing interest in the role of post-transcriptional regulation of gene expression by microRNAs (miRNAs) and their influence on cancer risk and clinical outcomes. miRNAs are small endogenous non-coding RNAs of ∼22–27 nucleotides. These pair with complementary binding sites located in the 3′-untranslated region (3′UTR) of messenger RNA (mRNA). Perfect complementation at the miRNA ‘seed’ site, the second to eighth nucleotide from the 5′-end of the miRNA, leads to mRNA degradation while imperfect pairing leads to inhibition of mRNA translation, the end result of both being post-transcriptional gene silencing (1–3). Based on the importance of seed pairing, multiple bioinformatics algorithms have been developed in order to predict miRNA-binding sites in mRNA sequences (4–8).
The efficiency of miRNA–mRNA pairing, and consequently gene expression levels, can be influenced by a number of factors, including miRNA expression levels, the presence of single-nucleotide polymorphisms (SNPs) in miRNA genes and the presence of SNPs located in miRNA-binding sites of mRNA 3′UTRs. Differential expression of miRNAs has been reported in various cancers, including those of bladder and breast, often linked to alterations in gene copy number of the miRNA genes (9,10). Polymorphisms in miRNA genes and miRNA-processing genes have also been found to be associated with the risk of cancers including breast cancer (11–13). Evolutionarily, Yu et al. (14) found that there was a negative selection against 3′UTR SNPs within predicted miRNA seed complementarity sites compared with other SNPs within the 3′UTR, due to their potentially deleterious effects. Using bioinformatics tools, studies have found SNPs in predicted miRNA-binding sites of candidate genes in breast and colorectal cancer that affected miRNA–mRNA binding and were associated with cancer risk (15,16).
Impaired DNA repair capacity due to DNA repair gene polymorphisms is associated with carcinogenesis and increased cancer risk, particularly for cancers where exposures to tobacco smoke and occupational carcinogens are major risk factors (17,18). Such carcinogens can cause a variety of DNA adducts including oxidative DNA damage some of which can lead to somatic mutations in genes, including oncogenes (19). The pathways involved in the repair of these DNA adducts are base excision repair (BER), nucleotide excision repair and double strand break (DSB) repair, mainly by non-homologous end joining (NHEJ) or homologous recombination.
Case–control studies have shown associations between DNA repair coding SNPs and bladder cancer susceptibility (20). The International Consortium on Bladder Cancer has performed a meta-analysis and pooled analyses of DNA repair gene coding SNPs on bladder cancer risk, confirming a small but significant association for three variants in ERCC2, NBS1 and XPC (21). In breast cancer, germ line non-synonymous mutations in the homologous recombination genes, BRCA1 and BRCA2, have long been linked with familial breast cancer (22) and studies have reported associations between SNPs in DSB repair with breast cancer susceptibility (23,24).
DNA repair is also involved in repairing DNA damage from chemotherapy and radiotherapy treatment of cancer cells. High ERCC1 mRNA and protein expression in bladder cancer cell lines are associated with increased sensitivity to cisplatin chemotherapy and ionizing radiation (25), while low bladder tumour MRE11 protein expression is associated with worse survival following radiotherapy treatment (26). However, the role of germ line coding DNA repair SNPs in predicting cancer survival following radiotherapy treatment has been less extensively investigated. Several studies have found associations of ATM, ERCC1 and XRCC1 SNPs with cancer-specific survival (CSS) in pancreatic, head and neck and lung cancer (27), whereas Sakano et al. (28) identified coding ERCC2 and XRCC1 SNPs potentially predictive of chemoradiotherapy outcomes in muscle-invasive bladder cancer patients.
We hypothesized that germ line SNPs in DNA repair genes could affect miRNA–mRNA binding and post-transcriptional regulation, thus influencing DNA repair capacity and cancer risk in normal somatic cells and radiotherapy sensitivity and outcome of cancer cells. To examine these hypotheses, we undertook a two-stage pilot study. First, we performed a bioinformatics analysis of 20 DNA repair genes, searching for SNPs residing in the 3′UTR, and examined their potential effects on miRNA-binding in silico. The seven candidate miRNA-binding SNPs which showed the greatest allele-specific differential binding were then genotyped in both a bladder cancer case–control series and also germ line DNA from patients with muscle invasive bladder cancer cases treated by radical radiotherapy. PARP1 rs8679 and RAD51 rs7180135 were found to be associated with bladder cancer risk and radiotherapy outcomes, respectively. Finally, the PARP1 SNP rs8679 was also assessed in a breast cancer case–control series.
Materials and methods
Study populations
Local ethical approval for the bladder cancer studies was obtained from the Leeds (East) Research Ethics Committee, and patients gave written informed consent. Recruitment of the bladder cancer case–control study population, comprising 752 bladder cancer cases and 704 controls, has been described previously (29). The radiotherapy cohort consisted of 202 muscle-invasive bladder cancer cases treated with external beam radiotherapy (52.5–55 Gy in 20 fractions over 4 weeks) between August 2002 and October 2009 at Cookridge Hospital and St James Institute of Oncology, Leeds, UK. A total of 257 breast cancer cases and 512 controls over the age of 30 years were recruited between February 1996 and April 2002. Cases were women treated in the Radiotherapy Department at Lyon-Sud Hospital, Pierre-Bénite, France, who gave written informed consent before participating in the study after local ethical committee approval had been obtained (30). Controls were age-matched female blood donors living in the vicinity of the hospital, obtained from community-based collections of the Regional Blood Transfusion Service. The health status of these controls was unknown, although any bias from such an individual having cancer would be expected to favour a null result.
Selection of polymorphisms
Twenty DNA repair genes involved in the BER, nucleotide excision repair, NHEJ, homologous recombination and DSB signalling pathways were selected for investigation (Supplementary Table I is available at Carcinogenesis Online). The methods for the bioinformatics prediction of putative miRNA-binding sites have been described previously (15). In July 2009, the 3′UTR genomic sequences for all 20 genes were obtained from the University of California Santa Cruz genome browser (http://genome.ucsc.edu, 3 January 2012, date last accessed) and putative miRNA-binding sites within these regions predicted using the miRBase (4), miRanda (5), Diana-MicroT (6), PicTar (7), TargetScanS (6) and MicroInspector (8) algorithms. Using the dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP, 3 January 2012, date last accessed), BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&BLAST_SPEC=SNP&BLAST_PROGRAMS=megaBlast&PAGE_TYPE=BlastSearch&SHOW_DEFAULTS=on&LINK_LOC=dbSNP_homepage, 3 January 2012, date last accessed) and BLAST-SNP (http://www.ncbi.nlm.nih.gov/SNP/snpblastByChr.html, 28th July 2009, date last accessed) algorithms, common SNPs [minor allele frequency (MAF) >0.10 for Caucasians] located within these putative miRNA-binding sites were identified. Using miRanda, the Gibbs free energy (ΔG, expressed in kilojoules per mole (kJ/mol)) for both the wild-type and variant alleles for each identified SNP was determined, with the difference in ΔG for the two alleles (wild-type allele ΔG − variant allele ΔG) calculated as ΔΔG. Therefore, negative ΔΔG values indicate weaker miRNA–mRNA binding in the presence of the variant allele than the wild-type allele (i.e. the wild-type allele ΔG is more negative), whereas positive ΔΔG values mean stronger binding in the presence of the variant allele (i.e. the variant allele ΔG is more negative). As more than one miRNA was predicted to bind to each SNP, thus theoretically increasing the likelihood of a true miRNA-binding site being present, the sum of all |ΔΔG|s >3 kJ/mol for each SNP (|ΔΔGtot|) was used to score the impact of that SNP on miRNA binding. The absolute values of ΔΔG (|ΔΔG|) were used to avoid positive and negative ΔΔG values negating each other. Of the top 10 SNPs with a |ΔΔGtot| >5 kJ/mol, two SNPs (NBS1 rs1063054 and XPC rs2229090) had previously been studied in the bladder cancer case–control study (29,31). The remaining eight SNPs were selected for genotyping.
Genotyping
Genomic DNA samples from the bladder cancer case–control population were sent for high-throughput Taqman genotyping to Gen-Probe Life Sciences in West Lothian, Scotland. Blind duplicates from 5% of samples were included to ensure concordance of genotyping calls. Pre-designed Taqman SNP genotyping assays (Applied Biosystems) were used for BRCA1 rs12516 and rs8176318, LIG3 rs4796030, NBS1 rs2735383, PARP1 rs8679 and RAD51 rs7180135. A custom Taqman assay was designed using the ABI assay by design tool for ATM rs227091 (forward primer: 5′-GGAGAAGAGGAAGAACAGAGGGATA-3′, reverse primer: 5′-GGACTAGAGGCTGCTGGAAAAG-3′, Reporter 1: VIC-5′-TCACTGCCAGTATCAG-3′, Reporter 2: FAM-5′-TCACTGCCATTATCAG-3′). Genotype calling analysis was performed using SDS 2.2 software (Applied Biosystems).
ATM SNP rs227091 also failed Taqman genotyping and was found to be located within Alu repeat regions, which was in turn immediately flanked by a LINE-1 repeat sequence. Therefore, polymerase chain reaction (PCR) primers within unique sequence flanking the repeat sequences were designed (forward: 5′-TGCACACAAGCCCATTCTTA-3′, reverse: 5′-AGCTGGGGGACAGAGAAATG-3′; Product 842 bp). PCR was carried out using HotStarTaq master mix (Qiagen) in 10 μl final volume using 40 pmol of each primer and 10 ng of genomic DNA. Reaction conditions were 95°C for 15 min, 36 cycles of 95°C, 30 s; 58°C 30 s; 72°C, 30 s, followed by 72°C for 10 min. DNA sequencing of PCR products, using primers 5′-GGAGTTTCGCTCTTGTCACC-3′ (rs1137918) and 5′-GTGCAGTGGCATGATCTCAG-3′ (rs227091), was carried out using standard dye terminator chemistry and data were collected using an Applied Biosystems Genetic Analyser 3130xl.
DNA sequencing was used to confirm the genotype of control samples. These control samples were run alongside PCR-amplified samples using the original Taqman custom assay (rs227091). There was complete concordance of genotype by sequencing and Taqman in all samples tested. One microlitre of 1:10 diluted PCR product was dried in a 384-well plate and resuspended in 2 μl of reaction mix containing 1× Taqman genotyping assay and 1× Taqman genotyping master mix (Applied Biosystems). Reaction conditions were 50°C for 2 min, 95°C for 10 min, then 40 cycles of 95°C, 15 s; 60°C, 60 s. Taqman fluorophore detection was carried out using an Applied Biosystems 7900 real-time PCR machine.
Another ATM SNP, rs1137918, failed genotyping by Taqman and sequencing, thus was not investigated further.
In the breast cancer case–control series, PARP1 rs8679 was genotyped using the above-mentioned pre-designed Taqman assay on 10 ng of genomic DNA with 2× Taqman genotyping master mix in a final volume of 10 μl. The genotyping PCR conditions were 95°C for 10 min, 50 cycles of 95°C, 15 s; 60°C, 60 s. The region around this SNP was sequenced in 10% of samples using 10 ng genomic DNA and 10 μM of the forward (5′-TGTGGGAAGACCAAAGGAAG-3′) and reverse (5′-ATAGAGAAGGCATCTGCATTTTTAAT-3′) primers in a final volume of 20 μl containing 1U of TaqPlatinum (Invitrogen), 0.2 mM final deoxyribonucleotide triphosphates and 1.5 mM final MgCl2. PCR conditions were 95°C for 3 min, 45 cycles of 95°C, 40 s; 58°C, 40 s; 72°C, 40 s, 72°C for 10 min. Five microlitres of the reaction volume was purified using 2 μl ExoSap (USB) and the DNA sequencing reaction was then carried out using standard dye terminator chemistry on an Applied Biosystems Genetic Analyser 3130.
Statistical analysis
Statistical analysis was performed using STATA software version 10 (StataCorp, TX) and SAS version 9.1 (SAS, CA). All SNPs were assessed for Hardy–Weinberg equilibrium of the SNP genotype frequency distribution in the controls, using a significance threshold of P = 0.05. In the bladder cancer case–control study, a multivariate logistic regression analysis was performed for the association of each SNP with bladder cancer risk estimating the odds ratio (OR) and 95% confidence interval (95% CI) and adjusting for gender, age (age at diagnosis for cases and age at consent for controls), smoking status (ever versus never) and occupational dye exposure (ever versus never). As the genetic inheritance model is unknown, a Cochran-Armitage trend test was deemed to provide the best power for detecting an association (32). The SNPs showing a significant risk effect were then incorporated into a secondary analysis for the total number of unfavourable genotype groups carried, adjusting for the same factors listed above. Logistic regression was used to assess the effect of PARP1 rs8679 in the breast cancer case–control series, the model was adjusted for age and the dominant gene effect was estimated.
In the muscle-invasive bladder cancer radiotherapy cohort, all SNPs were assessed as risk factors for CSS considering a dominant gene effect. Kaplan–Meier and Cox proportional hazard models were performed adjusting for age at diagnosis, gender, tumour (WHO stages 1–4) and nodal stage (WHO stages 0–2) and neoadjuvant chemotherapy (received versus not received). CSS was used as bladder cancer patients treated by radiotherapy tend to be elderly thus more likely to die of other causes (33).
With 752 cases and 704 controls, the bladder cancer case–control study had a statistical power of at least 80% for detecting an OR of 1.50 for a MAF of 0.2 at a 1% significance level. For 100 cancer-specific events (assuming an overall 50% 3 year CSS), the muscle-invasive bladder cancer cohort had a statistical power of 80% of detecting a hazard ratio of 2.00 in rare homozygotes and heterozygotes combined for a MAF of 0.2 at a 5% significance level.
Results
Bioinformatics predictions
Of the 20 genes, 5 genes, ATRX, CETN2, DNA-PK, RAD50 and XRCC1, had no SNPs located within their 3′UTR, whereas the 3′UTR SNPs in LIG1 and PCNA were not sited within a predicted miRNA-binding site. Of the remainder, there were 21 common SNPs (MAF > 0.10) located within a predicted miRNA-binding site in 13 genes (Supplementary Table II is available at Carcinogenesis Online). Ten of these SNPs had a |ΔΔGtot| >5 kJ/mol. Of these 10 SNPs, 2 had been previously genotyped in the bladder cancer case–control study, namely NBS1 rs1063054 (29) and XPC rs2229090 (31) and showed no association. XPC rs2229090 has also been imputed from Hapmap phase 2 variants in the Nijmegen and Icelandic bladder cancer study populations (B.Kiemeney, personal communication), with no association found with bladder cancer risk (combined OR = 0.99, 95% CI = 0.89–1.09, P = 0.80) despite this variant having the highest predicted |ΔΔGtot| of 113.27 for differential miRNA binding. The remaining eight candidate SNPs and corresponding predicted miRNA-binding sites with a |ΔΔG| >5 kJ/mol are listed in Table I.
Table I.
Candidate SNPs selected for genotyping in the case–control study and predicted miRNA binding with ΔΔG >3 kJ/mol
| Gene, dbSNP ID and allele substitution | MAF | Predicted miRNA binding | ΔΔGa | |ΔΔGtot|b |
| LIG3 rs4796030 C>A | 0.47 | miR-612 | −5.36 | 42.17 |
| miR-423-3p | −5.23 | |||
| miR-346 | −5.17 | |||
| miR-221 | −4.49 | |||
| miR-888* | −4.36 | |||
| miR-512-5p | −3.90 | |||
| miR-615-3p | −3.70 | |||
| miR-222 | −3.44 | |||
| miR-525-3p | −3.30 | |||
| miR-508-5p | −3.22 | |||
| ATM rs227091 T>C | 0.44 | miR-217 | −5.41 | 38.91 |
| miR-338-3p | −5.37 | |||
| miR-199b-5p | −4.79 | |||
| miR-199a-5p | −4.70 | |||
| miR-24 | −3.83 | |||
| miR-593* | −3.49 | |||
| miR-134 | −3.26 | |||
| miR-196a | 3.34 | |||
| miR-939 | 4.72 | |||
| BRCA1 rs12516 C>T | 0.36 | miR-874 | −9.92 | 25.76 |
| miR-324-3p | −8.83 | |||
| miR-623 | −7.01 | |||
| BRCA1 rs8176318 G>T | 0.35 | miR-328 | −4.10 | 21.82 |
| miR-565 | −3.95 | |||
| miR-149 | −3.71 | |||
| miR-146b-3p | −3.49 | |||
| miR-345 | −3.30 | |||
| miR-892b | −3.27 | |||
| PARP1 rs8679 T>C | 0.17 | miR-145 | −5.71 | 17.42 |
| miR-105 | −4.91 | |||
| miR-630 | −3.62 | |||
| miR-302a | 3.18 | |||
| ATM rs1137918 A>G | 0.22 | miR-615-5p | −5.14 | 12.80 |
| miR-193a-5p | −3.69 | |||
| miR-939 | 3.97 | |||
| NBN (NBS1) rs2735383 G>C | 0.32 | miR-499 | −4.36 | 8.70 |
| miR-508-3p | −4.34 | |||
| RAD51 rs7180135 A>G | 0.47 | miR-197 | 6.87 | 6.87 |
The ΔΔG was calculated by deducting the variant ΔG from the wild-type ΔG. A negative ΔΔG indicates decreased binding in the variant compared with the wild-type, whereas a positive ΔΔG indicates increased binding in the variant.
|ΔΔGtot| is the total of the absolute values of ΔΔG (|ΔΔG|).
Effects on bladder and breast cancer susceptibility
The bladder cancer and breast cancer study population demographics have been detailed previously (29,30). ATM rs227091 was successfully genotyped by Taqman genotyping of a nested PCR product for each sample, with full concordance in replicates and an assay failure rate of 6.25%. The remaining six candidate SNPs were genotyped with >98% concordance in replicates and an assay failure rate of <5%. Non-concordant replicates were due to exhaustion of these samples and so these were dropped from analysis. Hardy–Weinberg equilibrium for all SNPs was maintained in the controls. In a logistic regression model for bladder cancer risk including gender, age, smoking status and occupational dye exposure, smoking and occupational exposures were independent risk factors for bladder cancer (adjusted OR = 1.77, 95% CI = 1.36–2.30, P < 0.001 and adjusted OR = 1.36, 95% CI = 1.03–1.79, P = 0.03, respectively), with no effects seen for age or gender.
Adjusted logistic regression of the seven SNPs showed that only PARP1 rs8679 (Ptrend = 0.05) was associated with increased bladder cancer risk (Table II). PARP1 rs8679 was also assessed in the breast cancer case–control study but was not associated with increased breast cancer risk (Ptrend = 0.45) though CC homozygotes were noted to have a greater risk of developing breast cancer compared with TT (adjusted OR = 1.90, 95% CI = 1.05–3.43, p = 0.03, data not shown).
Table II.
Multivariate logistic regression analysis of candidate SNPs on bladder cancer risk
| Candidate SNP | Observed MAF (reported MAF) | Genotype | Case | Control | ORa (95% CI) | Ptrendb |
| LIG3 rs4796030 C>A | 0.46 (0.47) | CC | 204 | 226 | 1 | |
| AC | 369 | 321 | 1.28 (1.00–1.63) | 0.15 | ||
| AA | 160 | 148 | 1.21 (0.90–1.64) | |||
| ATM rs227091 T>C | 0.45 (0.44) | TT | 201 | 209 | 1 | |
| CT | 320 | 341 | 0.92 (0.69–1.21) | 0.70 | ||
| CC | 146 | 148 | 0.93 (0.69–1.27) | |||
| BRCA1 rs12516 C>T | 0.33 (0.36) | CC | 342 | 292 | 1 | |
| CT | 297 | 318 | 0.80 (0.64–1.01) | 0.43 | ||
| TT | 79 | 68 | 1.04 (0.72–1.50) | |||
| PARP1 rs8679 T>C | 0.21 (0.21) | TT | 412 | 424 | 1 | |
| CT | 266 | 214 | 1.29 (1.02–1.62) | 0.05 | ||
| CC | 45 | 37 | 1.23 (0.77–1.95) | |||
| BRCA1 rs8176318 G>T | 0.32 (0.35) | GG | 349 | 296 | 1 | |
| GT | 303 | 321 | 0.80 (0.64–1.00) | 0.29 | ||
| TT | 78 | 70 | 0.98 (0.68–1.41) | |||
| NBS1 rs2735383 G>C | 0.32 (0.32) | GG | 305 | 313 | 1 | |
| CG | 337 | 308 | 1.09 (0.87–1.36) | 0.27 | ||
| CC | 69 | 59 | 1.22 (0.83–1.81) | |||
| RAD51 rs7180135 A>G | 0.44 (0.47) | AA | 224 | 237 | 1 | |
| AG | 369 | 327 | 1.06 (0.80–1.41) | 0.97 | ||
| GG | 145 | 135 | 0.90 (0.66–1.21) |
Adjusted for age, gender, smoking status and occupational exposure.
Statistically significant P-values (P ≤ 0.05) are in bold.
Visual inspection of Table II suggests that the dominant model may better describe the gene effects of PARP1 rs8679. Under a dominant model (data not shown), PARP1 rs8679 (CT + CC genotypes) as well as LIG3 rs4796030 (AC + AA genotypes) were associated with increased bladder cancer risk (P = 0.03 and p = 0.05, respectively). The combined effects of LIG3 rs4796030 and PARP1 rs8679 was assessed by collapsing unfavourable genotypes (AC + AA and CT + CC genotypes, respectively) for analysis on bladder cancer risk (Table III). This demonstrated an additive genotype–dose effect with the number of unfavourable SNP genotype groups carried (Ptrend = 0.002). These results, however, should be interpreted with care in view of the potential application of the incorrect genetic model and the use of multiple comparisons.
Table III.
Multivariate logistic regression analysis of number of unfavourable LIG3 rs4796030 and PARP1 rs8679 SNP genotypes on bladder cancer risk
| Number of unfavourable genotypes carrieda | Case | Control | ORb (95% CI) | Ptrendc |
| 0 | 115 | 145 | 1 | 0.002 |
| 1 | 375 | 352 | 1.33 (0.99–1.78) | |
| 2 | 220 | 169 | 1.65 (1.20–2.29) |
Unfavourable genotypes defined as LIG3 rs4796030 AC + AA and PARP1 rs8679 CT + CC.
Adjusted for age, gender, smoking status and occupational exposure.
Statistically significant P-values (P ≤ 0.05) are in bold.
Effects on radiotherapy outcomes
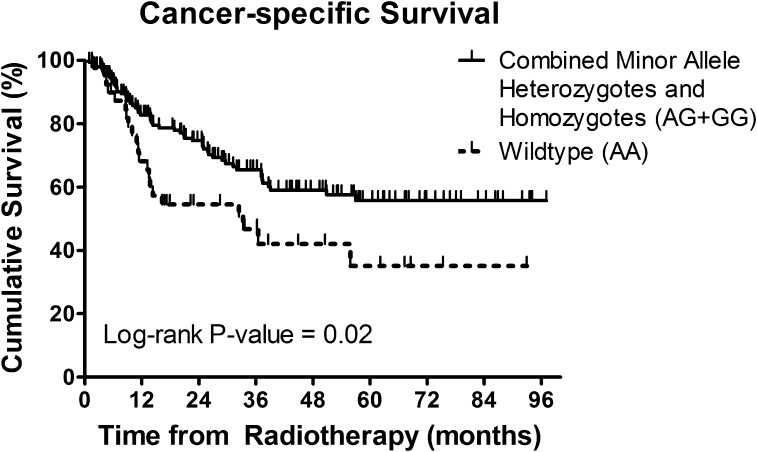
In the muscle-invasive bladder cancer cohort treated by radical radiotherapy, improved 5-year CSS rates were seen in carriers of at least one RAD51 rs7180135 minor allele (AG + GG) compared with common homozygotes (AA) (Figure 1; 56.2 versus 35.1%, respectively) in multivariate analysis (adjusted hazard ratio 0.52, 95% CI = 0.31–0.87, P = 0.01) (Table IV).
Fig. 1.
Kaplan–Meier survival curves of CSS in combined heterozygotes and rare homozygotes of RAD51 rs7180135 (AG + GG) versus wild-type (AA).
Table IV.
Cox multivariate analysis of CSS following radical radiotherapy and candidate SNP genotype using a dominant model
| Candidate SNP | Genotype | CSS |
||
| N | HR (95% CI)a | P-valueb | ||
| LIG3 rs4796030 C>A | CC | 56 | 1 | |
| AC + AA | 129 | 1.39 (0.81–2.39) | 0.23 | |
| ATM rs227091 T>C | TT | 52 | 1 | |
| CT + CC | 133 | 1.15 (0.62–2.17) | 0.65 | |
| BRCA1 rs12516 C>T | CC | 85 | 1 | |
| CT + TT | 101 | 0.84 (0.52–1.34) | 0.45 | |
| PARP1 rs8679 T>C | TT | 117 | ||
| CT + CC | 71 | 1.19 (0.74–1.92) | 0.46 | |
| BRCA1 rs8176318 G>T | GG | 87 | 1 | |
| GT + TT | 93 | 0.89 (0.55–1.42) | 0.62 | |
| NBS1 rs2735383 G>C | GG | 81 | 1 | |
| CG + CC | 101 | 0.95 (0.59–1.52) | 0.82 | |
| RAD51 rs7180135 A>G | AA | 50 | 1 | |
| AG + GG | 139 | 0.52 (0.31–0.87) | 0.01 | |
HR, hazard ratio.
Adjusted for age, gender, tumour and nodal stage and adjuvant chemotherapy.
Statistically significant P-values (P ≤ 0.05) are in bold.
Discussion
To our knowledge, this is the first study demonstrating the effects of DNA repair gene 3′UTR SNPs within putative miRNA-binding sites on bladder cancer susceptibility and radiotherapy outcomes. We found PARP1 rs8679 and possibly LIG3 rs4796030 to be associated with increased bladder cancer risk, with an additive effect seen with carriage of both SNPs. In a breast cancer case–control series, PARP1 rs8679 was also associated with increased breast cancer risk in individuals homozygous for the variant. The candidate SNPs were also investigated as potential predictive markers of radiotherapy treatment outcome in muscle-invasive bladder cancer, and we found that carriers of the RAD51 rs7180135 minor allele had improved 5-year CSS with an absolute survival advantage at 5 years of 21.1%.
LIG3 is a BER protein and is stabilized by XRCC1. It is recruited through interactions with PARP1 and XRCC1 to DNA SSBs and ligates the SSBs generated during short-patch BER. It may also provide a ‘back-up’ ligase for the long-patch BER subpathway (34). PARP1, another BER protein, carries out the poly(ADP-ribosyl)ation of histones, topoisomerases and DNA repair signalling proteins following DNA damage, resulting in chromatin unwinding and recruitment of DNA repair proteins (35). However, there is also evidence that PARP1 is involved in other DNA repair pathways and forms part of an alternative NHEJ repair pathway together with LIG3 and XRCC1 (36). The total ΔΔG (Table I) for the miRNAs predicted to bind to LIG3 rs4796030 and PARP1 rs8679 was lower for the variant alleles (A and C, respectively) compared with the wild-type alleles (C and T, respectively). Thus, the presence of the variant allele would be expected to be associated with a decrease in miRNA–mRNA binding and an increased LIG3 and PARP1 expression. This result is counter-intuitive as it might be expected that higher levels of DNA repair enzymes would be associated with increased repair and reduced cancer risk. However, the over-expression of DNA repair enzymes in tumours may provide a selective advantage for tumour cell growth and an over-expression has been associated with a greater risk of metastasis (37). The additive effect seen in our study of carrying both LIG3 rs4796030 and PARP1 rs8679 on bladder cancer risk suggests that either an enhanced BER pathway or the alternative NHEJ pathway (an error-prone DSB repair pathway compared with classical NHEJ) predisposes individuals to bladder cancer. PARP1 competes with Ku (classical NHEJ) for binding to DSB ends and initiating the alternative NHEJ pathway (38). Increased PARP1 and LIG3 expression due to LIG3 rs4796030 and PARP1 rs8679 could thus possibly promote DSB repair via alternative NHEJ over the classical pathway resulting in more error-prone repair and mutagenesis, with the greatest influence seen in carriers of both SNPs.
Of the published literature in bladder cancer on the miRNAs predicted to bind to LIG3 rs4796030, a pre-miRNA SNP of miR-423 has previously been associated with increased bladder cancer risk (39), whereas miR-221 has been reported to be upregulated in bladder cancer cell lines with miR-221 silencing leading to apoptosis (40). For the PARP1 rs8679 SNP, miR-145 binding was predicted to have the greatest ΔΔG (−5.71 kJ/mol), with the variant C allele ΔG being less negative than the wild-type T allele ΔG (Table I, a similar profile to that of two of the other three miRNAs predicted to bind this site) suggesting a decreased miR-145 binding to the variant PARP1 mRNA 3′UTR and thus increased PARP1 expression. Previous studies have reported down-regulation of miR-145 expression in bladder cancer (10,41), whereas transfection of miR-145 in bladder cancer cell lines has a tumour suppressor effect, reducing cell proliferation and inducing apoptotic pathways (42). Clearly, there is a need to assess LIG3 and PARP1 protein levels and miRNA expression profiles in breast and bladder tumours to determine the influence of these miRNAs on protein expression especially with current interest in the use of PARP1 inhibitors as a single agent or as a chemo- or radio-sensitizer in cancer treatment (35).
Several association studies have identified 3′UTR SNPs in candidate genes for their influence on cancer risk with significant SNPs then investigated for putative miRNA-binding sites (43,44). In bladder cancer, we previously found an association for the DSB repair MRE11 rs2155209 3′UTR SNP with increased bladder cancer risk (29). In the current study, this variant is predicted to be within the binding sites for miR-584 and miR-744 but the minor allele is predicted to alter the ΔΔG for either miRNA by <1 kJ/mol (Supplementary Table II is available at Carcinogenesis Online), thus the mechanism underlying this association remains to be determined. Several studies have used in silico methods to identify SNPs located within predicted miRNA-binding sites then evaluated their influence on cancer susceptibility (15,16,45). Using the bioinformatics analysis methods described above, Landi et al. (15) reported an association between 3′UTR SNPs in predicted miRNA-binding sites and cancer risk, showing that CD86 3′UTR SNP rs17281995 was predicted to affect the binding of four miRNAs and was associated with increased colorectal cancer risk (15). Chin et al. (45) demonstrated that KRAS 3′UTR SNP rs712 was sited within a let-7-binding site and was associated with increased lung cancer risk and also using a luciferase with KRAS 3′UTR reporter assay, resulted in increased luciferase activity compared with wild-type (45). Nicoloso et al. (16) also found two predicted miRNA-binding site SNPs located within the coding regions of BRCA1 and TGFR1 that affected post-transcriptional miRNA regulation (16), indicating possible miRNA-binding sites within the 5′UTR and coding regions of our candidate genes.
The role of miRNA-binding site polymorphisms as prognostic markers was highlighted by Brendle et al. (46). They identified a 3′UTR SNP within the ITGB4 gene associated with worse breast cancer survival and oestrogen receptor-negative tumours (46). As ionizing radiation from radiotherapy causes cancer cell death by inducing DNA damage, the candidate SNPs were thus investigated as potential predictive markers of radiotherapy treatment outcomes in muscle-invasive bladder cancer. We found the RAD51 rs7180135 G minor allele to be associated with improved survival. miR-197 is predicted to bind more strongly to the G allele (ΔΔG 6.87 kJ/mol) resulting in the reduction of RAD51 expression. RAD51 is involved in DSB homologous recombination repair; thus in tumours, alteration of RAD51 expression could potentiate tumour radiosensitization. Low tumour RAD51 mRNA expression has previously been associated with lower local recurrence and improved survival following adjuvant chemotherapy and radiotherapy treatment in breast cancer (47). Differential expression of miR-197 has been reported in several cancers with up-regulation seen in squamous cell carcinoma of the tongue and male breast carcinoma and down-regulation in gastric cancer (48–50). Plasma miR-197 levels have also been proposed as a potential diagnostic biomarker having been reported to be elevated in lung cancer (51). In human colon cancer cell lines, miR-197 expression levels were down-regulated following oxaliplatin and 5-fluorouracil treatment (52). In response to DNA damage, the effect of miR-197 down-regulation in cancer cells could thus be to increase DNA repair by increasing RAD51 expression. Our results suggest that the RAD51 rs7180135 genotype and its possible effects on miR-197-mRNA-binding may predict tumour radiosensitivity and radiotherapy outcomes in bladder cancer. If successfully validated, this might be used clinically as a predictive marker of radiotherapy outcome. Future studies of miR-197 as a therapeutic target to improve radiotherapy outcomes would also be worthwhile.
In this study, we have examined seven polymorphisms for associations, using multiple genetic models, with risk of bladder cancer and cause-specific mortality following radiotherapy. One of these polymorphisms was further tested for association with breast cancer risk. Given the complex correlation structure between these multiple tests, it is difficult to calculate the number of tests carried out in order to correct for multiple comparisons using a method such as the Bonferroni correction. If we simply correct for the number of SNPs genotyped, seven, the Bonferroni-corrected significance threshold would be 0.007. As such none of the associations observed would remain statistically significant. Similarly, the SNPs were all chosen due to their prior probability of functional significance, due to their influence on miRNA binding. However, the magnitude of this prior probability is also difficult to estimate for use in correction methods such as the false-positive report probability (53). Due to the subjectivity of the assumptions necessary for these corrections, we have chosen to not formally correct the P-values reported and thus acknowledge that the results should be interpreted with caution with the possibility of false-positive discovery.
In conclusion, we have examined the 3′UTR of 20 candidate DNA repair genes using in silico methods to identify SNPs that potentially affect miRNA binding. We have found associations between PARP1 rs8679 and increased bladder cancer and breast cancer risk and RAD51 rs7180135 and improved CSS following radiotherapy treatment in muscle-invasive bladder cancer. While the first of its kind in bladder cancer, our study also corroborates the work of others in other tumour types, investigating the association of miRNA-binding site SNPs and cancer susceptibility (15,16,45). If our findings can be successfully validated in further series and functional effects demonstrated in suitable assays, they may give further insight into the biology of bladder carcinogenesis, allow testing of the RAD51 SNP as a potential predictive biomarker and also reveal potential targets for new cancer treatments.
Supplementary material
Supplementary Tables 1 and II can be found at http://carcin.oxfordjournals.org/
Funding
Yorkshire Cancer Research (L350 to M.T.W.T.); Cancer Research UK (C15140/A11505 to A.E.K.); Inserm and Institut Curie (to Inserm U612).
Supplementary Material
Acknowledgments
We thank Helen Snowden in processing the bladder cancer DNA samples for genotyping and Olga Sinilnikova for allowing access to a panel of DNA samples collected from healthy blood donors.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- BER
base excision repair
- CI
confidence interval
- CSS
cancer-specific survival
- DSB
double strand break
- MAF
minor allele frequency
- mRNA
messenger RNAs
- miRNA
microRNAs
- NHEJ
non-homologous end joining
- PCR
polymerase chain reaction
- SNP
single-nucleotide polymorphism
- 3′UTR
3′-untranslated region
References
- 1.Liu Z, et al. MicroRNA: an emerging therapeutic target and intervention tool. Int. J. Mol. Sci. 2008;9:978–999. doi: 10.3390/ijms9060978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeng Y, et al. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl Acad. Sci. USA. 2003;100:9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brennecke J, et al. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.John B, et al. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krek A, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 6.Lewis BP, et al. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 7.Rusinov V, et al. MicroInspector: a web tool for detection of miRNA binding sites in an RNA sequence. Nucleic Acids Res. 2005;33:W696–W700. doi: 10.1093/nar/gki364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiriakidou M, et al. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc. Natl Acad. Sci. USA. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dyrskjot L, et al. Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009;69:4851–4860. doi: 10.1158/0008-5472.CAN-08-4043. [DOI] [PubMed] [Google Scholar]
- 11.Horikawa Y, et al. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin. Cancer Res. 2008;14:7956–7962. doi: 10.1158/1078-0432.CCR-08-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Z, et al. Genetic variants in selected pre-microRNA genes and the risk of squamous cell carcinoma of the head and neck. Cancer. 2010;116:4753–4760. doi: 10.1002/cncr.25323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang R, et al. A genetic variant in the pre-miR-27a oncogene is associated with a reduced familial breast cancer risk. Breast Cancer Res. Treat. 2010;121:693–702. doi: 10.1007/s10549-009-0633-5. [DOI] [PubMed] [Google Scholar]
- 14.Yu Z, et al. Aberrant allele frequencies of the SNPs located in microRNA target sites are potentially associated with human cancers. Nucleic Acids Res. 2007;35:4535–4541. doi: 10.1093/nar/gkm480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Landi D, et al. Polymorphisms within micro-RNA-binding sites and risk of sporadic colorectal cancer. Carcinogenesis. 2008;29:579–584. doi: 10.1093/carcin/bgm304. [DOI] [PubMed] [Google Scholar]
- 16.Nicoloso MS, et al. Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility. Cancer Res. 2010;70:2789–2798. doi: 10.1158/0008-5472.CAN-09-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berwick M, et al. Markers of DNA repair and susceptibility to cancer in humans: an epidemiologic review. J. Natl Cancer Inst. 2000;92:874–897. doi: 10.1093/jnci/92.11.874. [DOI] [PubMed] [Google Scholar]
- 18.Goode EL, et al. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol. Biomarkers Prev. 2002;11:1513–1530. [PubMed] [Google Scholar]
- 19.Wu X, et al. Genetic susceptibility to tobacco-related cancer. Oncogene. 2004;23:6500–6523. doi: 10.1038/sj.onc.1207811. [DOI] [PubMed] [Google Scholar]
- 20.Kiltie AE. Molecular epidemiology of DNA repair genes in bladder cancer. Methods Mol. Biol. 2009;472:281–306. doi: 10.1007/978-1-60327-492-0_12. [DOI] [PubMed] [Google Scholar]
- 21.Stern MC, et al. Polymorphisms in DNA repair genes, smoking, and bladder cancer risk: findings from the international consortium of bladder cancer. Cancer Res. 2009;69:6857–6864. doi: 10.1158/0008-5472.CAN-09-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tonin P, et al. Frequency of recurrent BRCA1 and BRCA2 mutations in Ashkenazi Jewish breast cancer families. Nat. Med. 1996;2:1179–1183. doi: 10.1038/nm1196-1179. [DOI] [PubMed] [Google Scholar]
- 23.Silva SN, et al. Breast cancer risk and common single nucleotide polymorphisms in homologous recombination DNA repair pathway genes XRCC2, XRCC3, NBS1 and RAD51. Cancer Epidemiol. 2010;34:85–92. doi: 10.1016/j.canep.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 24.Thomas G, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p.11.2 and 14q24.1 (RAD51L1) Nat. Genet. 2009;41:579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawashima A, et al. Excision repair cross-complementing group 1 may predict the efficacy of chemoradiation therapy for muscle-invasive bladder cancer. Clin. Cancer Res. 2011;17:2561–2569. doi: 10.1158/1078-0432.CCR-10-1963. [DOI] [PubMed] [Google Scholar]
- 26.Choudhury A, et al. MRE11 expression is predictive of cause-specific survival following radical radiotherapy for muscle-invasive bladder cancer. Cancer Res. 2010;70:7017–7026. doi: 10.1158/0008-5472.CAN-10-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parliament MB, et al. Single nucleotide polymorphisms of DNA repair genes as predictors of radioresponse. Semin. Radiat. Oncol. 2010;20:232–240. doi: 10.1016/j.semradonc.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 28.Sakano S, et al. Single nucleotide polymorphisms in DNA repair genes might be prognostic factors in muscle-invasive bladder cancer patients treated with chemoradiotherapy. Br. J. Cancer. 2006;95:561–570. doi: 10.1038/sj.bjc.6603290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choudhury A, et al. Analysis of variants in DNA damage signalling genes in bladder cancer. BMC Med. Genet. 2008;9:69. doi: 10.1186/1471-2350-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moullan N, et al. Polymorphisms in the DNA repair gene XRCC1, breast cancer risk, and response to radiotherapy. Cancer Epidemiol. Biomarkers Prev. 2003;12:1168–1174. [PubMed] [Google Scholar]
- 31.Sak SC, et al. Comprehensive analysis of 22 XPC polymorphisms and bladder cancer risk. Cancer Epidemiol. Biomarkers Prev. 2006;15:2537–2541. doi: 10.1158/1055-9965.EPI-06-0288. [DOI] [PubMed] [Google Scholar]
- 32.Li Q, et al. Robust tests for single-marker analysis in case-control genetic association studies. Ann. Hum. Genet. 2009;73:245–252. doi: 10.1111/j.1469-1809.2009.00506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kotwal S, et al. Similar treatment outcomes for radical cystectomy and radical radiotherapy in invasive bladder cancer treated at a United Kingdom specialist treatment center. Int. J. Radiat. Oncol. Biol. Phys. 2008;70:456–463. doi: 10.1016/j.ijrobp.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 34.Petermann E, et al. Roles of DNA ligase III and XRCC1 in regulating the switch between short patch and long patch BER. DNA Repair (Amst) 2006;5:544–555. doi: 10.1016/j.dnarep.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 35.Megnin-Chanet F, et al. Targeting poly(ADP-ribose) polymerase activity for cancer therapy. Cell. Mol. Life Sci. 2010;67:3649–3662. doi: 10.1007/s00018-010-0490-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Audebert M, et al. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 2004;279:55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 37.Sarasin A, et al. Overexpression of DNA repair genes is associated with metastasis: a new hypothesis. Mutat. Res. 2008;659:49–55. doi: 10.1016/j.mrrev.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 38.Wang M, et al. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang H, et al. Evaluation of genetic variants in microRNA-related genes and risk of bladder cancer. Cancer Res. 2008;68:2530–2537. doi: 10.1158/0008-5472.CAN-07-5991. [DOI] [PubMed] [Google Scholar]
- 40.Lu Q, et al. MicroRNA-221 silencing predisposed human bladder cancer cells to undergo apoptosis induced by TRAIL. Urol. Oncol. 2010;28:635–641. doi: 10.1016/j.urolonc.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 41.Catto JW, et al. Distinct microRNA alterations characterize high- and low-grade bladder cancer. Cancer Res. 2009;69:8472–8481. doi: 10.1158/0008-5472.CAN-09-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ostenfeld MS, et al. miR-145 induces caspase-dependent and -independent cell death in urothelial cancer cell lines with targeting of an expression signature present in Ta bladder tumors. Oncogene. 2010;29:1073–1084. doi: 10.1038/onc.2009.395. [DOI] [PubMed] [Google Scholar]
- 43.Pharoah PD, et al. The role of KRAS rs61764370 in invasive epithelial ovarian cancer: implications for clinical testing. Clin. Cancer Res. 2011;17:3742–3750. doi: 10.1158/1078-0432.CCR-10-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang D, et al. Genetic variants in MicroRNA biosynthesis pathways and binding sites modify ovarian cancer risk, survival, and treatment response. Cancer Res. 2010;70:9765–9776. doi: 10.1158/0008-5472.CAN-10-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chin LJ, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3' untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brendle A, et al. Polymorphisms in predicted microRNA-binding sites in integrin genes and breast cancer: ITGB4 as prognostic marker. Carcinogenesis. 2008;29:1394–1399. doi: 10.1093/carcin/bgn126. [DOI] [PubMed] [Google Scholar]
- 47.Le Scodan R, et al. DNA repair gene expression and risk of locoregional relapse in breast cancer patients. Int. J. Radiat. Oncol. Biol. Phys. 2010;78:328–336. doi: 10.1016/j.ijrobp.2009.07.1735. [DOI] [PubMed] [Google Scholar]
- 48.Wong TS, et al. Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin. Cancer Res. 2008;14:2588–2592. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- 49.Lehmann U, et al. Identification of differentially expressed microRNAs in human male breast cancer. BMC Cancer. 2010;10:109. doi: 10.1186/1471-2407-10-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X, et al. miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol. Cancer Res. 2011;9:824–833. doi: 10.1158/1541-7786.MCR-10-0529. [DOI] [PubMed] [Google Scholar]
- 51.Zheng D, et al. Plasma microRNAs as novel biomarkers for early detection of lung cancer. Int. J. Clin. Exp. Pathol. 2011;4:575–586. [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou J, et al. 5-Fluorouracil and oxaliplatin modify the expression profiles of microRNAs in human colon cancer cells in vitro. Oncol. Rep. 2010;23:121–128. [PubMed] [Google Scholar]
- 53.Wacholder S, et al. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J. Natl Cancer Inst. 2004;96:434–442. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.