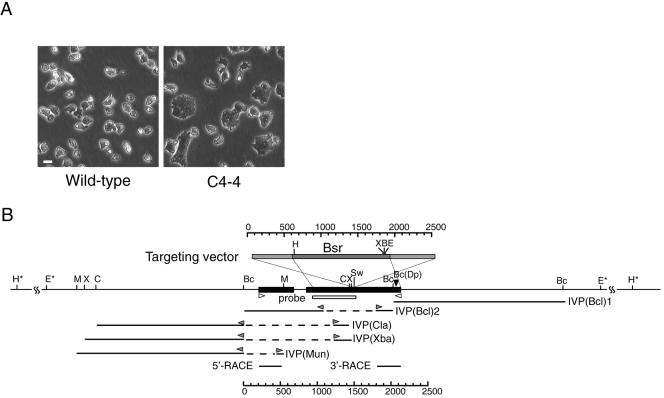
Figure 1.
C4-4 REMI-mutant (A) and schematic representation of the gene disrupted in C4-4 (B), which we named dwwA. (A) Phase contrast micrographs of clone C4--4 and wild-type cells grown on plastic dishes. (B) Restriction map around dwwA and the targeting vector. The targeting vector (top box) used to knockout dwwA has an insertion of the Bsr cassette (dark gray box) at the unique SwaI site, and extends to both sides of the SwaI site (light gray boxes) in exon 2. The two exons are shown as black boxes. The underlines indicate five genomic DNA fragments cloned from the C4-4 cells by inverse PCR. Bc(Dp) indicates the insertion site of the original REMI-mutant. White box indicates a region used as a probe for Southern blotting. Two white arrowheads indicate the primers used for RT-PCR. Gray arrowheads indicate the inverse PCR primers used for genome walking. Restriction enzyme sites: B, BamHI; Bc, BclII; C, ClaI; E, EcoRI; H, HindIII; M, MunI; Sw, SwaI; X, XbaI; and Dp, DpnII. Asterisks indicate restriction enzyme sites that were predicted by Southern blotting.