Fig. 1. Strategy for quantification of differential protein expression using QCET.
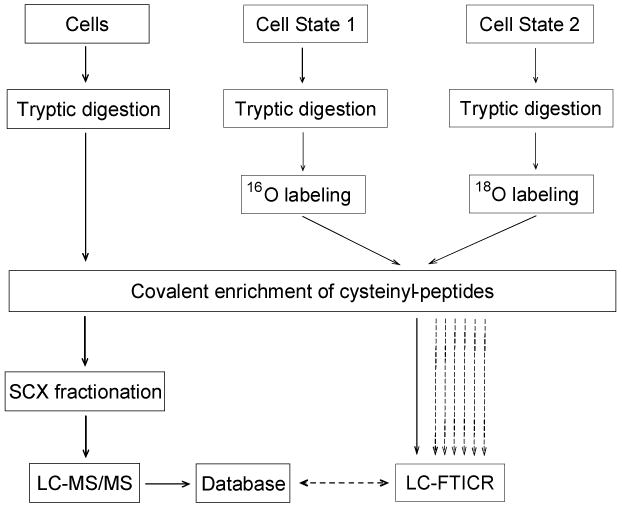
In the first stage, proteins from different cell states are mixed and digested by trypsin, followed by cysteinyl-peptide enrichment using thiol-affinity resin. The enriched cysteinyl-peptides are fractionated by SCX chromatography with each fraction analyzed by LC-MS/MS. An AMT tag database is generated based on the calculated masses and normalized elution times for all identified peptides. In the second stage, the two protein mixtures representing two different cell states are digested by trypsin separately. The resulting tryptic peptides are labeled by trypsin-catalyzed oxygen exchange using 16O- and 18O-enriched water, respectively. The two samples are combined and cysteinyl-peptides are selectively enriched and analyzed by LC-FTICR without pre-fractionation. Peptide features are identified and quantified by matching to the AMT tag database without the need for additional LC-MS/MS analyses. Once an AMT tag database is established for a biological system, the system can be extensively investigated in a high throughput manner by analyzing samples generated under different conditions using LC-FTICR. (Modified from ref. 6 with permission from the American Chemical Society.)
