Abstract
OBJECTIVE
The NOD mouse strain has been widely used to investigate the pathology and genetic susceptibility for type 1 diabetes. Induced pluripotent stem cells (iPSCs) derived from this unique mouse strain would enable new strategies for investigating type 1 diabetes pathogenesis and potential therapeutic targets. The objective of this study was to determine whether somatic fibroblasts from NOD mice could be reprogrammed to become iPSCs, providing an alternative source of stem cells for the production of genetically modified NOD cells and mice.
RESEARCH DESIGN AND METHODS
Adult tail-tip fibroblasts from male NOD mice were reprogrammed by retroviral transduction of the coding sequences of three transcription factors, OCT4, SOX2, and KLF4, in combination with a histone deacetylase inhibitor, valproic acid.
RESULTS
Eighteen NOD iPSC lines were generated, and three of these cell lines were further characterized. All three cell lines exhibited silencing of the three reprogramming transgenes and reactivation of endogenous pluripotent markers (OCT4, SOX2, NANOG, REX1, and SSEA1). These NOD iPSCs readily differentiated in vitro to form embryoid bodies and in vivo by teratoma formation in immunodeficient mice. Moreover, NOD iPSCs were successfully transfected with a reporter transgene and were capable of contributing to the inner cell mass of C57BL/6 blastocysts, leading to the generation of a chimeric mouse.
CONCLUSIONS
Adult tail-tip fibroblasts from NOD mice can be reprogrammed, without constitutive ectopic expression of transcription factors, to produce iPSCs that exhibit classic mouse embryonic stem cell (ESC) features. These NOD iPSCs can be maintained and propagated under normal ESC culture conditions to produce genetically altered cell lines, differentiated cells, and chimeric mice.
NOD mice spontaneously develop type 1 diabetes, similar to humans with this disease, and provide a valuable model for investigating type 1 diabetes pathogenesis under controlled genetic and environmental settings (1). The derivation of pluripotent embryonic stem cells (ESCs) from the NOD strain has proven elusive until a recent study identified conditions for derivation and maintenance by using small molecules that inhibit the key differentiation-inducing pathways (2–7).
ESC-like cells can also be generated by forced expression of defined transcription factors in somatic cells, with the resulting cells termed “induced pluripotent stem cells” (iPSCs) (8–12). Because of their pluripotent nature, iPSCs are amenable to gene targeting via homologous recombination and germline competency (9,13,14), making them a promising alternative source of pluripotent cells for producing genetically modified tissues or mice from strains for which derivation of conventional ESCs has proven difficult.
In a previous study, Hanna et al. (4) claimed the constitutive ectopic expression of KLF4 or c-MYC was an absolute requirement for derivation and maintenance of NOD iPSCs. By contrast, we observed a combination of retroviruses encoding OCT4, SOX2, and KLF4 (OSK), despite being silenced (15), are capable of fully reprogramming adult NOD mouse tail-tip fibroblasts (nTTFs) to generate NOD iPSCs.
RESEARCH DESIGN AND METHODS
Mice.
Mice, including NOD/Lt mice for tail-tip fibroblasts isolation, were maintained in a specific pathogen-free animal facility under Monash University approved ethics (MMCA2007/34).
Tail-tip fibroblast isolation.
An ∼1.5 cm length of tail-tip from 8-week-old male NOD mice was washed with 70% ethanol and PBS, the superficial dermis was peeled away, and the remaining tissue was cut into 1-mm pieces using a scalpel. Five or six pieces were plated in one well of a six-well plate and cultured with 2 mL of nTTF medium (Dulbecco’s modified Eagle’s medium with 10% FBS and 50 units/mL penicillin/streptomycin) for 5–7 days. Cells migrating out were trypsinized and expanded into T25 flasks (passage 1).
Retrovirus production and iPSC induction.
Retrovirus production and iPSC induction were performed as described previously with minor modifications (16). Briefly, pMX-based retroviral vectors (Addgene) encoding mouse OCT4, SOX2, and KLF4 sequences were independently transfected into Plat-E cells. The next day, nTTFs at passage 4 were seeded at a density of 1 × 105 cells per well in six-well plates overnight. The virus-containing supernatants were collected 48 h after transfection, mixed, filtered, and added to the nTTFs together with 4 µg/mL polybrene (Sigma-Aldrich, St. Louis, MO) for 24 h. Subsequently, nTTFs were cultured in mouse ESC medium supplemented with 2 μmol/L valproic acid (VPA), which was refreshed daily for 7 days.
Cell culture and colony pick-up.
Mouse ESC culture medium was composed of knockout-Dulbecco’s modified Eagle’s medium, 20% knockout serum replacement, 0.1 mmol/L nonessential amino acids, 1 mmol/L Glutamax, 0.1 mmol/L β-mercaptoethanol (all from Invitrogen, Carlsbad, CA), and 1,000 units/mL leukemia inhibitory factor (LIF) (Millipore, Billerica, MA). Cells were cultured in a 37°C, 5% CO2 incubator. At day 14 after transduction, colonies were picked individually and incubated with 20 µL of 0.25% trypsin/1 mmol/L EDTA (Invitrogen) for 5 min at 37°C in a well. The colonies were dissociated into single cells and transferred to wells in 96-well plates with mouse embryonic fibroblast (MEF) feeder cells and 200 µL ESC medium. The iPSCs were further expanded in six-well plates.
Immunocytochemistry.
Immunocytochemical staining was performed as described previously (16). Primary antibodies were OCT4 (1:100, Santa Cruz Biotechnology, Inc., Santa Cruz, CA), NANOG (1:100, Abcam, Cambridge, U.K.), and SSEA-1 (1:100, Millipore).
RT-PCR and genomic DNA PCR.
Total RNA was isolated using the RNeasy kit (QIAGEN, Hilden, Germany), and genomic DNA contamination was removed using DNA-free kit (Ambion, Austin, TX). One microgram of total RNA was used for cDNA synthesis using Superscript III Reverse Transcriptase and Oligo (dT) primers (Invitrogen).
Genomic DNA was isolated from approximately 2 × 106 iPSCs for each cell line using DNeasy Blood and Tissue kit (QIAGEN). PCR was performed using primers listed in Table 1.
TABLE 1.
Primer sequences
| Gene name | Forward primers (5′ to 3′) | Reverse primers (5′ to 3′) | |
|---|---|---|---|
| For Tg genomic PCR and RT-PCR | |||
| Endogenous | Oct4 | TCTTTCCACCAGGCCCCCGGCTC | TGCGGGCGGACATGGGGAGATCC |
| Sox-2 | TAGAGCTAGACTCCGGGCGATGA | TTGCCTTAAACAAGACCACGAAA | |
| Klf4 | GCGAACTCACACAGGCGAGAAACC | TCGCTTCCTCTTCCTCCGACACA | |
| Nanog | TCAAGGACAGGTTTCAGAAGCA | GCTGGGATACTCCACTGGTG | |
| Rex1 | GGACTAAGAGCTGGGACACG | GCTGCTTCCTTCTTGAACAAT | |
| β-actin | GGAATCCTGTGGCATCCATGAAAC | AAAACGCAGCTCAGTAACAGTCCG | |
| Transgene | Oct4 | TTGGGCTAGAGAAGGATGTGGTTC | TTATCGTCGACCACTGTGCTGCTG |
| Sox2 | GGTTACCTCTTCCTCCCACTCCAG | TTATCGTCGACCACTGTGCTGCTG | |
| Klf4 | GCGAACTCACACAGGCGAGAAACC | TTATCGTCGACCACTGTGCTGCTG | |
| For bisulfite-sequencing nested PCR | |||
| Oct4 | 1st round | TTGAGGAGTGGTTTTAGAAATAATTGGTAT | CCCAACCCTACTCCAACCCTACTA |
| 2nd round | GGGTAAGTAAGAATTGAGGAGTGGTTT | CCCAACCCTACTCCAACCCTACTA | |
| Nanog | 1st round | AAGTATGGATTAATTTATTAAGGTAGTT | AAAAAACCCACACTCATATCAATATA |
| 2nd round | AAGTATGGATTAATTTATTAAGGTAGTT | CAACCAAATCAACCTATCTAAAAA | |
| For the three germ layer marker RT-PCR | |||
| Differentiation markers | Nestin | TCTGGAAGTCAACAGAGGTGG | ACGGAGTCTTGTTCACCTGC |
| Vimentin | GATGTTTCCAAGCCTGACCTC | GGCGTTCCAGAGACTCGTTAG | |
| GATA6 | CTGAATACTTGAGGTCACTGTTCTCGGG | ACCTTATGGCGTAGAAATGCTGAGGGTG | |
| Brachyury | CATGTACTCTTTCTTGCTGG | GGTCTCGGGAAAGCAGTGGC | |
| FoxA2 | TGGTCACTGGGGACAAGGGAA | GCAACAACAGCAATAGAGAAC | |
| Sox17 | TTTGTGTATAAGCCCGAGATGG | AAGATTGAGAAAACACGCATGAC | |
Bisulfite genomic sequencing.
Genomic DNA from NOD iPS, ES (D3 line), and nTTF cells were extracted and processed for bisulfite genomic sequencing analysis as described previously (17). Nested-PCR and sequencing was performed using primers listed in Table 1.
Differentiation of NOD-iPSCs.
To form embryoid bodies (EBs) in vitro, NOD-iPSCs were dissociated into single cells and resuspended in mES medium without LIF in bacteriological Petri dishes (Falcon, BD Biosciences) for 1 week. To form teratomas in vivo, ∼2 × 106 NOD iPSCs were injected into the hind leg muscle of NOD/SCID mice. Teratoma tissue was dissected out 4 weeks postinjection and processed for hematoxylin–eosin staining by the Monash Institute of Medical Research Histology Laboratory core facility.
Blastocyst injection and embryo transfer.
Host embryos were collected at the blastocyst stage from C57BL/6 female mice at 3.5 days post coitum, and 10–15 NOD iPSCs were injected into each. After a recovery period of ∼2 h, 10 to 15 injected blastocysts were transferred into one uterine horn of pseudopregnant CBA×C57BL/6 F1 female mice at 2.5 days post coitum. Chimeric mice were identified by coat color, and the male chimera was assessed for germline transmission by mating with NOD female mice.
Flow cytometric analysis of splenic B cells.
Single cell suspensions were prepared from individual spleens and erythrocytes were lysed by treating the sample with tris-NH4Cl followed by washing with FACS buffer containing 2% FBS. FC receptor binding was blocked by incubating cells with 2.4G2 (BD Biosciences Pharmingen, San Diego, CA). The following monoclonal antibodies were used in multiparameter flow cytometric analysis: anti-CD19 (6D5, Alexa Fluor 700 conjugated) and anti-H-2Kb (AF6–88.5, PerCP/Cy5.5 conjugated) from Biolegend and anti-H-2Kd (SF1–1.1, fluorescein isothiocyanate conjugated), anti-I-Ag7 (OX-6, fluorescein isothiocyanate conjugated), and anti-I-Ab (AF6–120.1, phycoerythrin conjugated) from BD Pharmingen. Cells were analyzed with an LSR Fortessa (BD Biosciences, Franklin Lakes, NJ) and FlowJo (Tree Star) software using propidium iodide incorporation to exclude dead cells.
Microsatellite genotyping of cell lines.
DNA was extracted from the NOD-iPS#5 cell line and genotyped for 112 microsatellite markers across the mouse genome covering 19 autosomes and the X chromosome. All markers used were polymorphic between NOD/Lt and C57BL/6 mouse strains. Genotyping was performed by the Australian Genome Research Facility Ltd., using the Applied Biosystems (Carlsbad, CA) 3730 DNA Analyzer and AB Gene Mapper Software.
RESULTS
During the 7 days of VPA treatment (Fig. 1A), clusters of cells started to aggregate together. By day 14 postviral infection, ES-like colonies were individually picked, trypsinized, and transferred to MEF feeder cells in 96-well plates for expansion (Fig. 1B). A total of 41 putative colonies were picked in three replicated experiments, and 18 colonies were successfully expanded in six-well plates (Supplementary Table 1). The cells formed compacted colonies showing a refractile halo effect when observed by phase-contrast microscopy (Fig. 1B), which is similar to that observed for mouse ESC colonies. The cells were routinely split in a 1:6 ratio every 2–3 days. The NOD-iPSCs have been maintained for >50 passages. Next, we transfected one of the NOD-iPSC lines with an mCherry expression cassette (CMV-mCherry-hygro) by LipofectaMINE (Invitrogen) as described previously (18). After selection with hygromycin and expansion, a stably transfected NOD-iPS-mCherry cell line was established (Fig. 1B).
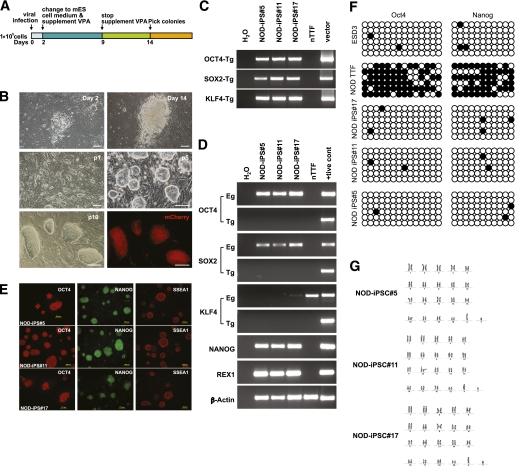
FIG. 1.
Generation of iPSCs from nTTFs. A: Schematic representation of the experimental protocol. nTTFs were plated at a density of 1 × 105 cells per well in six-well plates and infected with retroviruses encoding OSK factors for 24 h. The infected nTTFs were cultured in fibroblast medium for 2 days, switched to mouse ESC medium supplemented with VPA for 1 week, and subsequently cultured in mouse ESC medium. Emerging iPSC colonies were picked ∼14 days after infection. B: Representative images of colonies observed at different stages as indicated in the panels (p, passage number) and the representative colonies stably transfected with CMV-mCherry-Hygro construct. C: Integration of OCT4, SOX2, and KLF4 transgenes in the three OSK-induced iPSC lines was confirmed by genomic PCR using transgene-specific primers, with H2O and nTTF as negative controls and the construct plasmids as positive control. D: Expression of endogenous genes OCT4, SOX2, KLF4, NANOG, and REX1; expression of transgenes OCT4, SOX2, and KLF4 in the tail-tip fibroblasts; and the three OSK-induced iPSC lines were assessed by RT-PCR, with H2O as negative control and appropriate samples as corresponding positive controls. β-Actin was used as a loading control. E: Immunofluorescence staining shows expression of pluripotency markers (OCT4, NANGO, and SSEA1) in the three OSK-induced iPSC lines. F: Methylation analysis of OCT4 and NANOG promoter regions. Genomic DNA from mouse ESCs (ESD3), fibroblasts (nTTF), and the three OSK-induced iPSC lines were processed for bisulfite sequencing. Each horizontal row of circles represents an individual sequencing result from one amplicon. Open and filled circles indicate unmethylated and methylated CpG dinucleotides, respectively. G: Karyotype of the three OSK-induced iPSC lines. (A high-quality digital representation of this figure is available in the online issue.)
Of the 18 NOD iPSC lines generated, three were selected for further characterization. We confirmed by genomic DNA PCR that the three reprogramming transgenes (OSK) were integrated in the genome of the iPSCs (Fig. 1C). RT-PCR analysis confirmed that OSK transgenes were efficiently silenced in the three NOD iPSC lines (Fig. 1D). In addition, reactivation of endogenous OCT4 and SOX2 genes, as well as other pluripotent-associated transcription factors such as NANOG and REX1 (also known as ZFP42), was observed in all NOD iPSC lines tested in the study (Fig. 1D). Immunohistochemical analysis showed that the iPSCs stained positively for OCT4, NANOG, and SSEA1 (Fig. 1E). Further, bisulfite sequencing analysis revealed that promoter regions examined for both OCT4 and NANOG were demethylated in NOD-iPSCs, in a manner similar to mouse ESCs (ESD3 cell line), compared with heavily methylated patterns observed for the same regions in the parental nTTF cells (Fig. 1F), suggesting epigenetic reprogramming to pluripotency. Karyotyping analysis showed a normal chromosome number (40, XY) in the three iPSC lines (Fig. 1G), and genome-wide genotyping (∼112 microsatellite markers) of NOD-iPS #5 cell line confirmed the NOD genetic background (Supplementary Table 2).
We next evaluated the differentiation potential of the NOD iPSCs in vitro by EB formation and in vivo by teratoma induction. The iPSCs readily formed EBs on culture in the absence of feeders and LIF (Fig. 2A). RT-PCR of the EBs showed strong suppression of pluripotency genes including OCT4 and NANOG, with coincident activation of lineage-specific genes representing the three germ layers: nestin (ectoderm); vimentin and brachyury (mesoderm); and Gata6, Sox17, and FoxA2 (endoderm) (Fig. 2B). All three NOD-iPSC lines generated well-differentiated teratomas in the NOD/SCID mice. Hematoxylin-eosin staining of teratoma sections showed tissues representative of the germ layers, including keratinized-epithelium (ectoderm), cartilage (mesoderm), and secreting-gland epithelium (endoderm) (Fig. 2C). Immunohistochemical staining of teratomas confirmed differentiation to the three germ layers (Supplementary Fig. 1).
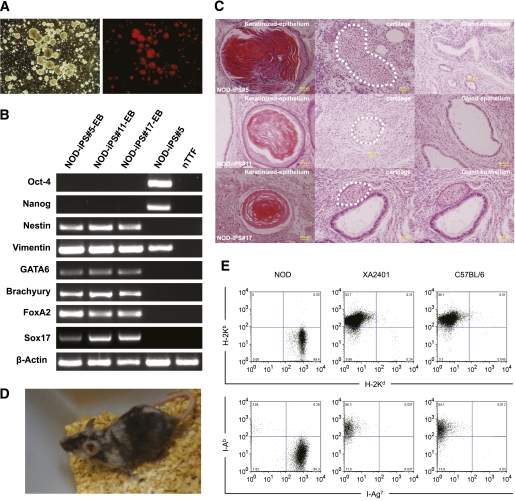
FIG. 2.
Differentiation of NOD iPSCs and chimera analysis. A: In vitro differentiation of CMV-mCherry-Hygro vector transfected NOD iPSCs to EBs (bright field and fluorescence field). B: RT-PCR analysis of total RNA isolated from EBs generated from the three NOD iPSC lines, one NOD iPSC line (No. 5), and nTTF. The expression of Oct4 and Nanog (pluripotency markers); vimentin and brachyury (mesoderm); Gata6, FoxA2, Sox17 (endoderm markers); and nestin (ectoderm marker) were examined. β-Actin was used as a loading control. C: In vivo differentiation of the three OSK-induced NOD iPSC lines. Histologic analysis of teratomas indicates iPSCs contribute to tissues from the three germ layers, including keratinized-epithelium (ectoderm), cartilage (mesoderm), and secreting-gland epithelium (endoderm). Tissues outlined by the dashed line are cartilage tissue. D: Contribution of NOD-iPS#5 cells in chimeric mouse (XA2401) as detected by agouti coat color. E: Hematopoietic chimerism assessment by flow cytometry analysis of spleen samples from a wild-type NOD mouse, the chimeric mouse (XA2401), and a C57BL/6 mouse. Samples were stained for and gated on CD19+ cells to identify B cells. Top: H-2Kb (C57BL/6) and H-2Kd (NOD) MHC class I expression on CD19+ cells. Bottom: I-Ab (C57BL/6) and I-Ag7 (NOD) MHC class II expression on CD19+ cells. (A high-quality digital representation of this figure is available in the online issue.)
A total of 24 C57BL/6 blastocysts injected with NOD-iPS#5 cells were transferred into the uteri of two pseudopregnant mice. Both recipients became pregnant and delivered a total of 17 pups, with one male pup (designated as XA2401) showing 40–50% coat color chimerism (Fig. 2D). We crossed the chimeric male with wild-type NOD female mice to examine germ-line transmission capability; however, no albino offspring resulted from three litters examined (18 pups born).
Last, we assessed the hematopoietic chimerism of male XA2401 by cell surface marker expressions of the major histocompatibility complex (MHC). NOD mice express the MHC class I molecule Kd and class II molecule I-Ag7, whereas C57BL/6 mice express Kb and I-Ab molecules, respectively. Flow cytometric analysis of B-cell expression of MHC molecules in the spleen indicated that the XA2401 mouse showed a C57BL/6 hematopoietic system (Fig. 2E), despite a variegated coat color chimerism.
DISCUSSION
The major goal of our current research was to demonstrate that somatic fibroblasts from NOD mice can be reprogrammed to generate iPSCs. We used three Yamanaka factors, OCT4, SOX2, and KLF4, but not c-MYC, to fulfill the reprogramming process of nTTF in this study. c-MYC was excluded because previous studies have shown that potential reactivation of the c-MYC transgene could result in tumor development in the chimeras and their progeny derived from the iPSCs (9,19). Instead, the chromatin remodeling small-molecule VPA (a histone deacetylase inhibitor) was used because it has been shown to greatly increase the efficiency of reprogramming human and mouse fibroblasts (20,21). Male (XY) donor cells were also chosen for induction to pluripotency for three reasons. 1) Male pluripotent cell-derived differentiated cells are easier to track in vivo, based on Y-chromosome specific probes, when used for transplantation studies into female recipients. 2) Aberrant X-inactivation associated with female (XX) ESC lines can result in XO cells, which compromises euploidy of cells, chimera generation, and germ-line transmission (22,23). 3) Male ESC chimeras can be rapidly bred with more than one wild-type female to produce multiple litters.
By using this modified approach, we show that NOD iPSCs are readily generated from nTTFs by transduction with retroviral vectors encoding OCT4, SOX2, and KLF4 and supplementation with VPA in the culture medium for 1 week. All three transgenes were silenced in the NOD iPSCs, suggesting complete reprogramming of the somatic nTTF. The NOD iPSCs could be dissociated into single cells by trypsinization and maintained for >50 passages. The NOD iPSCs derived in this study maintained ESC-like pluripotent characteristics and did not convert to an epiblast stem (EpiS) cell-like state, which is characterized by distinct colonies with flat morphology (24,25). This contrasts with the findings of Hanna et al. (4), because their NOD iPSCs appeared to be metastable and acquired an alternative EpiS cell-like identity after removal of exogenous KLF4 or cMyc factor expression. The EpiS cell colonies not only were morphologically distinct from mouse ESCs but also lost the ability to be propagated after dissociation into single cells by trypsinization. In our study, the derivation and maintenance of NOD iPSCs were achieved without constitutive expression of KLF4 transgene or addition of small molecules of GSK3/ERK pathway inhibitors. This difference may be due to the source of adult somatic tissue: we used nTTF, whereas Hanna et al. (4) used NOD MEF. Nonetheless, our modified method provides a robust alternative strategy for the generation of ESC-like pluripotent stem cells from a nonpermissive mouse strain without using embryos.
ACKNOWLEDGMENTS
This work was funded in part by financial support from the Australian Phenomics Network, which is an initiative of the Australian Federal Government’s National Collaborative Research Infrastructure Strategy; the Australian National Health and Medical Research Council (575552); and the National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases (5R01-DK-062882). This work was supported by the Victorian Government’s Operational Infrastructure Support Program. M.K.O’B. is a National Health and Medical Research Council Fellow (No. 545805).
No potential conflicts of interest relevant to this article were reported.
J.L. conducted the study and wrote the article. M.P.A. and H.S. conducted the study and reviewed the article. M.K.O’B., T.C.B., and P.J.V. directed the study and reviewed the article.
The authors thank Susan Chapman from the Australian Phenomics Network for blastocyst microinjection and embryo transfer and Vijesh Vaghjiani at the Centre for Reproduction and Development, Monash Institute of Medical Research, for assistance with immunohistochemistry.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db10-1540/-/DC1.
REFERENCES
- 1.Atkinson MA, Leiter EH. The NOD mouse model of type 1 diabetes: as good as it gets? Nat Med 1999;5:601–604 [DOI] [PubMed] [Google Scholar]
- 2.Nagafuchi S, Katsuta H, Kogawa K, et al. Establishment of an embryonic stem (ES) cell line derived from a non-obese diabetic (NOD) mouse: in vivo differentiation into lymphocytes and potential for germ line transmission. FEBS Lett 1999;455:101–104 [DOI] [PubMed] [Google Scholar]
- 3.Ying QL, Wray J, Nichols J, et al. The ground state of embryonic stem cell self-renewal. Nature 2008;453:519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanna J, Markoulaki S, Mitalipova M, et al. Metastable pluripotent states in NOD-mouse-derived ESCs. Cell Stem Cell 2009;4:513–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nichols J, Jones K, Phillips JM, et al. Validated germline-competent embryonic stem cell lines from nonobese diabetic mice. Nat Med 2009;15:814–818 [DOI] [PubMed] [Google Scholar]
- 6.Ohta H, Ohinata Y, Ikawa M, et al. Male germline and embryonic stem cell lines from NOD mice: efficient derivation of GS cells from a nonpermissive strain for ES cell derivation. Biol Reprod 2009;81:1147–1153 [DOI] [PubMed] [Google Scholar]
- 7.Yang W, Wei W, Shi C, et al. Pluripotin combined with leukemia inhibitory factor greatly promotes the derivation of embryonic stem cell lines from refractory strains. Stem Cells 2009;27:383–389 [DOI] [PubMed] [Google Scholar]
- 8.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006;126:663–676 [DOI] [PubMed] [Google Scholar]
- 9.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature 2007;448:313–317 [DOI] [PubMed] [Google Scholar]
- 10.Takahashi K, Okita K, Nakagawa M, Yamanaka S. Induction of pluripotent stem cells from fibroblast cultures. Nat Protoc 2007;2:3081–3089 [DOI] [PubMed] [Google Scholar]
- 11.Brambrink T, Foreman R, Welstead GG, et al. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell 2008;2:151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yusa K, Rad R, Takeda J, Bradley A. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat Methods 2009;6:363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wernig M, Zhao JP, Pruszak J, et al. Neurons derived from reprogrammed fibroblasts functionally integrate into the fetal brain and improve symptoms of rats with Parkinson’s disease. Proc Natl Acad Sci U S A 2008;105:5856–5861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanna J, Wernig M, Markoulaki S, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science 2007;318:1920–1923 [DOI] [PubMed] [Google Scholar]
- 15.Jähner D, Stuhlmann H, Stewart CL, et al. De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature 1982;298:623–628 [DOI] [PubMed] [Google Scholar]
- 16.Liu J, Sumer H, Leung J, et al. Late passage human fibroblasts induced to pluripotency are capable of directed neuronal differentiation. Cell Transplant. 2010 August 17 [Epub ahead of print] [DOI] [PubMed]
- 17.Tat PA, Sumer H, Jones KL, Upton K, Verma PJ. The efficient generation of induced pluripotent stem (iPS) cells from adult mouse adipose tissue-derived and neural stem cells. Cell Transplant 2010;19:525–536 [DOI] [PubMed]
- 18.Liu J, Jones KL, Sumer H, Verma PJ. Stable transgene expression in human embryonic stem cells after simple chemical transfection. Mol Reprod Dev 2009;76:580–586 [DOI] [PubMed] [Google Scholar]
- 19.Nakagawa M, Koyanagi M, Tanabe K, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol 2008;26:101–106 [DOI] [PubMed] [Google Scholar]
- 20.Huangfu D, Osafune K, Maehr R, et al. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol 2008;26:1269–1275 [DOI] [PubMed] [Google Scholar]
- 21.Huangfu D, Maehr R, Guo W, et al. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol 2008;26:795–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robertson EJ, Evans MJ, Kaufman MH. X-chromosome instability in pluripotential stem cell lines derived from parthenogenetic embryos. J Embryol Exp Morphol 1983;74:297–309 [PubMed] [Google Scholar]
- 23.Zvetkova I, Apedaile A, Ramsahoye B, et al. Global hypomethylation of the genome in XX embryonic stem cells. Nat Genet 2005;37:1274–1279 [DOI] [PubMed] [Google Scholar]
- 24.Tesar PJ, Chenoweth JG, Brook FA, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 2007;448:196–199 [DOI] [PubMed] [Google Scholar]
- 25.Brons IG, Smithers LE, Trotter MW, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 2007;448:191–195 [DOI] [PubMed] [Google Scholar]