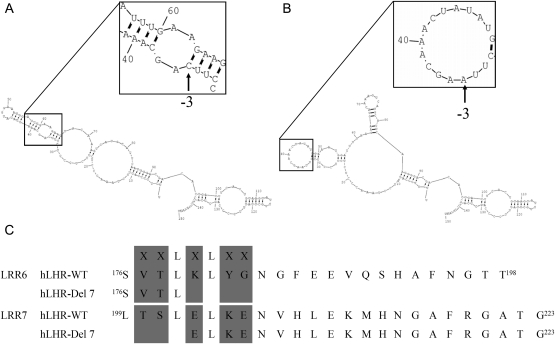
Figure 1.
Prediction of secondary RNA structure of LHR. (A) Secondary RNA structure of wild-type LHR. The nucleotide sequence (150 bp) covering 37 bp 5′ and 44 bp 3′ of the exon was analyzed by the program RNAstructure. Regions covering the 3′ acceptor splice site were given in the inset at a higher magnification. IVS6-3 acceptor splice sites are indicated by arrows. (B) Secondary RNA structure of mutant LHR. The free energy (△G) calculated by RNAstructure program was not significantly different than that of the wild-type transcript. (C) LRR6 and LRR7 of LHR-WT and LHR-Del7. LHR-Del7 caused an in-frame deletion of 23 amino acids. The XXLXLXX motif constituted the hydrophobic core for hormone binding. LHR-Del7 destroyed the hydrophobic core in LRR6 and LRR7. (The concave surface of the LRR region was shown in gray).