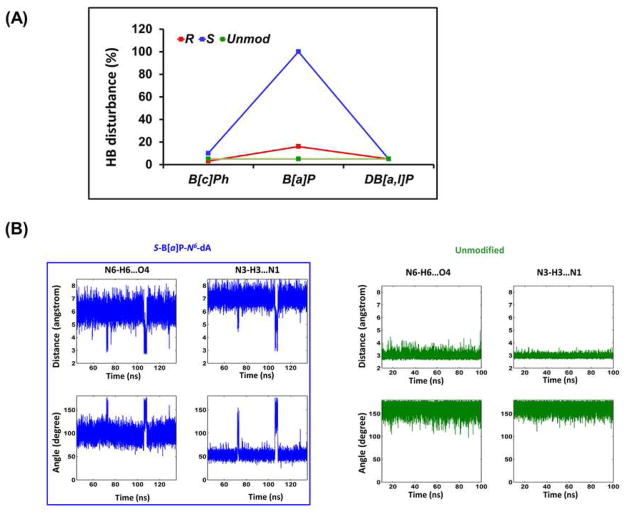
Figure 3.
Watson-Crick hydrogen bonding at the A6*:T17 base pair featuring the complete disruption in the S-B[a]P-N 6-dA adduct case. (A) Percent disruption of hydrogen bonds at A6*:T17. The values for the most disrupted of the two hydrogen bonds in the Watson-Crick base pair are shown. The full data set for the central 3-mer is given in Table S3A, Supporting Information. Our hydrogen bond quality index reveals the same pattern of distortions (Table S3B, Supporting Information). (B) Time dependence of hydrogen bond distances and angles for the A6*:T17 in the S-B[a]P-N 6-dA adduct, and at A6:T17 for the unmodified control, showing disrupted and dynamic distances and angles at the A6*:T17 base pair. The time dependent fluctuations of hydrogen bond distances and angles for all adducts are shown in Figure S3, Supporting Information; these reveal that all are similar to the unmodified control, except for the S-B[a]P-N 6-dA adduct.