Abstract
Purpose: This study investigates the feasibility of T2* to be a diagnostic indicator of early breast cancer in a mouse model. T2* is sensitive to susceptibility effects due to local inhomogeneity of the magnetic field, e.g., caused by hemosiderin or deoxyhemoglobin. In these mouse models, unlike in patients, the characteristics of single mammary ducts containing pure intraductal cancer can be evaluated.
Methods: The C3(1)SV40Tag mouse model of breast cancer (n = 11) and normal FVB/N mice (n = 6) were used to measure T2* of normal mammary gland tissue, intraepithelial neoplasia, invasive cancers, mammary lymph nodes, and muscle. MRI experiments were performed on a 9.4T animal scanner. High resolution (117 microns) axial 2D multislice gradient echo images with fat suppression were acquired first to identify inguinal mammary gland. Then a multislice multigradient echo pulse sequence with and without fat suppression were performed over the inguinal mammary gland. The modulus of a complex double exponential decay detected by the multigradient echo sequence was used to fit the absolute proton free induction decay averaged over a region of interest to determine the T2* of water and fat signals.
Results: The measured T2* values of tumor and muscle are similar (∼15 ms), and almost twice that of lymph nodes (∼8 ms). There was a statistically significant difference (p < 0.03) between T2* in normal mammary tissue (13.7 ± 2.9 ms) and intraductal cancers (11 ± 2.0 ms) when a fat suppression pulse was applied.
Conclusions: These are the first reported T2* measurements from single mammary ducts. The results demonstrated that T2* measurements may have utility for identifying early pre-invasive cancers in mouse models. This may inspire similar research for patients using T2* for diagnostic imaging of early breast cancer.
Keywords: mouse models of cancer, T2*, breast cancer, MRI
INTRODUCTION
MRI plays a major role in the diagnosis and management of breast cancer. Early and accurate detection of breast cancer decreases morbidity and mortality and significantly reduces the financial cost of treatment.1 While MRI reliably detects invasive cancers—there is still a need to improve detection and characterization of early cancers, especially ductal carcinoma in situ (DCIS). Contrast enhanced imaging, combined with T1- and T2-weighted imaging are the mainstays of clinical breast MRI. T2*-weighted imaging is an alternative approach that does not require contrast injection. T2*-weighted imaging is sensitive to the highly deoxygenated blood that is often found in tumor blood vessels, due to the high magnetic susceptibility of deoxyhemoglobin.2 This is commonly referred to as blood oxygen level dependent (BOLD) contrast.2 Quantitative measurement of T2* provides an alternative approach to assessment of early stage breast cancers. T2*-weighted imaging is not used in routine clinical practice to assess breast lesions but may provide clinically useful information—particularly for patients who cannot tolerate contrast media injection. T2* value may also be useful in preclinical studies as a marker for initiation and progression of mammary cancer. T2*-weighted imaging is sensitive to deoxygenated tumor blood vessels,3 microcalcifcations, and other sources of local magnetic susceptibility gradients, and thus may be a useful marker for early intraductal cancers. Here, we report an initial step toward testing the diagnostic utility of quantitative T2* values for breast/mammary cancer.
To evaluate whether T2* value can identify and stage mammary cancers—we measured T2* in mammary glands of the C3(1) SV40Tag transgenic mouse. This is a widely used model for spontaneous human breast cancer and is especially helpful for modeling early cancers.4, 5 These mice develop mammary intraepithelial neoplasia (MIN) that resembles DCIS in women. The intraductal cancer frequently progresses to the invasive stage. The mice develop atypical ductal hyperplasia at ∼8 weeks of age, MIN at ∼12 weeks of age, and invasive tumors at ∼16 weeks of age.4 Therefore, this model allows imaging of cancers at various stages.
This model is particularly advantageous for studies of early intraductal cancers, because individual ducts containing neoplasia can be reliably imaged.5, 6 This is difficult in humans because single ducts containing pure DCIS can rarely be resolved in clinical MR images. Typical spatial resolution of clinical MRI is 1 mm in-plane with 2 mm thickness, and the spatial resolution is even lower when contrast enhanced MRI data are acquired with high temporal resolution. Much higher resolution is required to visualize pure DCIS.7 Thus, the results reported here are the first measurements of T2* in single mammary ducts with pure intraductal cancer.
Measurements of T2* in mammary glands, whether murine or human, pose special problems. Like women’s breasts, mouse mammary glands contain fat that complicates the MR signal and makes accurate T2* measurements more difficult. High spectral and spatial resolution (HiSS) MR imaging has been used to separate water and fat spectra for detection of breast cancers,8, 9 and images of T2* in murine mammary cancer were calculated from the water resonance in each voxel.10, 11 Alternatively, the T2* can be measured by analyzing a train of gradient echoes in the time domain. This approach is particularly useful when the proton free induction decay (FID) is sampled unevenly and/or for a relatively short period of time. In this case, the T2* values can be extracted by fitting the time domain signal intensity as a function of echo time using an exponential decay model. A mono-exponential model is used for tissue with almost no fat. If tissue contains both water and fat as is the case for mammary glands, then a modulus of a complex double exponential decay function can be used in order to accurately fit the curve.12 This method has been demonstrated by O’Reagan et al to derive relative signal from water and fat in the liver, and results obtained from time domain analysis were verified by comparison with proton MR spectroscopy.
In this study, the inguinal mammary glands of mice were imaged on a 9.4 T. The T2* values for normal mammary gland tissue, MIN invasive cancers, mammary lymph nodes, and muscle were measured for C3(1) SV40Tag mice. For comparison, the T2* values for normal mammary gland tissue, mammary lymph nodes, and muscle in FVB/N wild-type mice were also measured. The T2* values were compared between normal mammary tissue and intraductal cancers to test whether it could be used in diagnosing early breast cancer. These are the first reported T2* values for pure intraductal cancer.
METHODS
Animals
All procedures were carried out in accordance with the University of Chicago Animal Care and Use Committee approval. A total of 17 mice were used in this study, including eleven C3(1) SV40 large T antigen (Tag) transgenic mice (age = 12–17 weeks) and six FVB/N wild-type mice (age = 15–17 weeks). Animals were anesthetized prior to imaging experiments, and anesthesia was maintained during imaging with 1.5% isofluorane. The temperature, heart rate, and respiration rate were monitored by SA Instruments (Stony Brook, NY), and the respiration rate was used to obtain gated images.
MRI experiments
The measurements were performed using a 9.4T Bruker scanner (Bruker-Biospin, Billerica, MA) with an open birdcage coil (3 cm length × 3 cm width × 2 cm height). High resolution axial gradient recalled echo images with fat suppression and respiratory gating (TR/TE = 675/7 ms, flip angle = 30°, field of view = 30 × 30 mm, matrix size = 256 × 256, number of slices = 41, slice thickness = 0.5 mm, number of excitation = 2) were acquired first to locate the inguinal mammary gland. Then a respiratory gated axial multislice and multigradient echo pulse sequence with bipolar read-out gradients was used with and without fat suppression with TR of ∼1000 ms (min TE = 1.5 ms, 9 echoes at 3 ms spacing, array size = 128 × 128, field of view = 30 × 30 mm, slice thickness = 1 mm, number of excitation = 1). Fat suppression was performed using a selective 90° pulse at −3.5 ppm relative to water which was followed by a spoiling gradient. To maintain the constant gating, the anesthesia vaporizer was quickly adjusted slightly if necessary based on mouse’s respiration rate.
Immediately following imaging, mice were euthanized, the mammary glands excised and fixed in 10% formalin over night. Normal mammary gland tissue, MIN, invasive cancers, and mammary lymph nodes were identified by MRI, and verified in hematoxylin and eosin (H&E) stained sections (5-μm thick H&E sections every 50 μm) by a pathologist, who has over 20 years of experience in the evaluation of breast and mouse mammary glands. H&E sections were imaged using an Olympus MVX10 and photographed at 12.6× magnification.
Data analysis
Data were analyzed and processed using in-house programs written in IDL (Research Systems, Inc., Boulder, CO). Since the proton free induction decay was sampled with only 9 echoes, and the “dwell time” for the first echo was considerably shorter than for subsequent echoes, this data acquisition scheme complicated analysis of the data in the spectral domain, and thus, data were analyzed in the time domain. Regions of interest (ROI) were selected and signal intensity was averaged over all pixels prior to fitting for calculation of T2*. For tissues with a fat signal that was less than 10% of the water signal, T2* was calculated by fitting the modulus MRI signal intensity [S(t)] as a function of echo time (i.e., the proton free induction decay), to the single exponential decay function: S(t)= Ae−t/T2*, where A is amplitude, t is echo time in ms. However, for tissues known to contain fat (fat signal greater than 10% of the water signal), ROIs were fit with the modulus of a complex double exponential decay function12
| (1) |
where, Aw and Af are the amplitude for water and fat components, respectively; T2*w and T2*f are the T2* values for water and fat, respectively; and Δω is the frequency difference and is fitted from the data. This function was used for fat suppressed data, as the fat suppression was incomplete, and the residual fat signal was greater than 10% of the water signal.
Due to a small sample size, the following nonparametric tests were performed in this study. A Wilcoxon matched-pairs signed-rank test was used to determine significance when comparing the same set of data with and without fat suppression. In order to determine the statistical significance of differences between T2* values in the various tissues, e.g., normal mammary gland tissue vs MIN, a Wilcoxon test was performed.
RESULTS
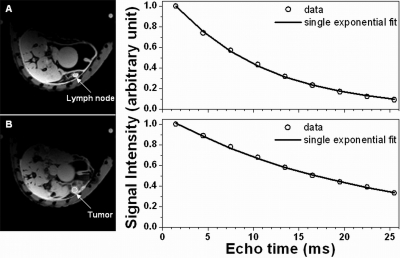
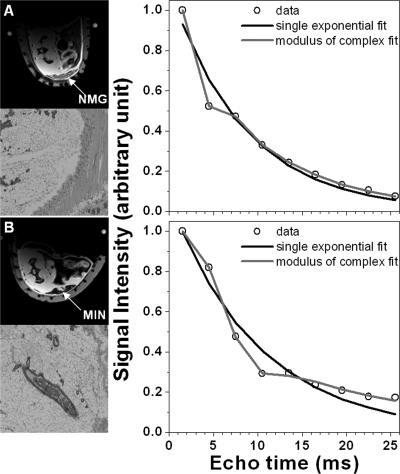
Both MRI and histopathology showed in situ cancer (MIN) and invasive tumors in the SV40Tag mice. No mammary cancers were found in wild-type mice. Of the 17 transgenic mice imaged, 8 had MIN lesions, and 5 had invasive tumors. Figure 1 shows a typical example of the single exponential decay model fitting proton free induction decays from lymph node and tumor ROI’s in SV40Tag mice. As expected, for ROIs in muscle (not shown here), lymph node, and tumor, the T2* curves are fit accurately by a single exponential decay function (average goodness of fitting parameter R2 = 0.994 ± 0.011). However, for normal mammary tissue [Fig. 2a] and MIN [Fig. 2b] ROIs, the modulus of complex double exponential decay model was required to fit the curve accurately, and account for the interference between fat and water signals. This is demonstrated by the improvement in average R2 value for complex double exponential fits (R2 = 0.993 ± 0.008) compared to single exponential fits (R2 = 0.944 ± 0.015). This was true even when fat suppression was used, since there was a significant residual fat signal. The difference between R2 = 0.993 and R2 = 0.944 is small, but it implies that the complex double exponential decay function fits the periodic oscillations of the signal decay curves more accurately (Fig. 2) than the single exponential.
Figure 1.
Selected axial images of transgenic mice (left column) and corresponding proton free induction decays (right column, data points shown by open circles) over the tissues indicated by the arrows for: (a) Lymph node tissue, and (b) tumor. Both lymph node and tumor FIDs can be accurately fit with a single exponential decay function (line).
Figure 2.
Selected axial fat suppressed images of mammary glands from transgenic mice (left column top) and corresponding T2* curves (right column, data given by open circles) over the ROIs indicated by the arrow for: (a) normal mammary gland (NMG) tissue and (b) MIN lesion. H&E sections (left column bottom) are provided at 12.6× magnification. Normal mammary tissue and MIN data are fit by both single (dark line) and complex double (light line) exponential decay functions. The modulus of the complex double exponential decay function (light line) is clearly necessary to accurately model the FIDs that include a fat component.
T2* values calculated using the single exponential decay model are reported in Table TABLE I. for the following tissues: tumor in SV40Tag mice, as well as lymph node and muscle from both SV40Tag and FVB/N. Use of fat suppression did not change the calculated T2* values for these tissues (paired Wilcox test p = 0.81, 0.19, 0.64, respectively). T2* values of tumor and muscle are almost the same, and almost twice that of lymph nodes. There was no difference for T2* values between SV40 Tag and wild-type mice in muscle (p = 0.18) and lymph node (p = 0.79). The fat T2* (measured without fat suppression) in normal mammary tissue was 3.6 ± 1.3 ms in transgenic and 5.0 ± 1.6 ms in wild-type mice.
TABLE I.
Average T2* (ms) values for tissue without fat signal calculated using a single exponential decay fit over ROIs of muscle and lymph node tissues for both SV40Tag transgenic mice and FVB/N normal mice, and calculated average T2* (ms) values over ROIs of tumor for SV40Tag transgenic mice only.
| Invasive tumor (n = 5) | Lymph node (n = 10) | Muscle (n = 11) | |
|---|---|---|---|
| SV40Tag with fat suppression | 14.6 ± 4.2 | 7.82 ± 2.1 | 14.9 ± 3.0 |
| SV40Tag | 14.9 ± 3.5 | 8.51 ± 3.1 | 15.5 ± 3.0 |
| FVB/N (n = 6) | N/A | 7.79 ± 2.3 | 12.6 ± 3.2 |
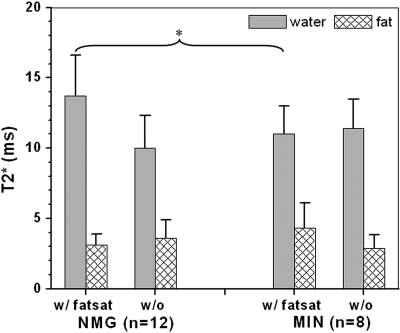
Figure 3 shows the water and fat T2* values calculated using the complex modulus double exponential decay model for normal mammary gland and MIN in SV40Tag mice. For normal mammary glands, a statistically significant difference was observed between calculated water T2* values measured with (13.7 ± 2.9 ms) and without (10.0 ± 2.3 ms) fat suppression (p < 0.005). For MIN, the calculated water T2* was almost the same with (11.0 ± 2.0 ms) and without (11.4 ± 2.1 ms) fat suppression. With fat suppression, T2* values in normal mammary glands are significantly longer (p < 0.03) than in MIN.
Figure 3.
T2* values are reported for MIN and normal mammary glands for both water and fat signals acquired with and without fat suppression. There is a significant difference between the water T2* in normal glands with and without fat suppression, with fat suppressed normal mammary glands having a longer T2*. When fat suppression is used, the calculated T2* is significantly longer for normal mammary glands than for MIN lesions.
Finally, in wild-type FVB/N NMG, without fat suppression measured water and fat T2* values are 9.4 ± 1.9 ms and 5.0 ± 1.6 ms, respectively. These values are similar to normal mammary gland T2* values measured in SV40Tag without fat suppression.
DISCUSSION
Use of a mathematical model to separate water and fat signals in the time domain was effective for accurate measurements of T2* in both transgenic and FVB/N mice. The modulus of a complex double exponential decay model provided excellent fits to experimental data, and separated the fat and water signals acquired from fat-rich murine mammary tissues. Even when a fat suppression pulse was applied, it was necessary to use the complex modulus of a double exponential decay model to fit the data due to a considerable residual fat signal. While the methods used here yielded promising results, other approaches to T2* measurements, such as the DIXON method, are available and should be evaluated. The advantage of the DIXON method is that it does not require curve fitting, and this facilitates pixel-by-pixel analysis. However, the DIXON method is commonly used when only two or three gradient echoes are acquired, and there can be significant errors in the calculation of the width of the water resonance or T2*.
When data were acquired with fat suppression, the measured T2* in MIN was significantly shorter than the T2* in normal tissue. The shorter T2* could be due to deoxygenated tumor neovasculature, or magnetic susceptibility effects of intraductal cancer cells and microcalcifications. Shorter T2* can also be produced by magnetic susceptibility gradients at the interface between fat and water—and the geometry of this interface may be different in MIN vs normal mammary glands. Although the MIN lesions were very small, typically ∼250 μm in diameter, the high spatial resolution of the images allowed effective segmentation of MIN, with minimal contamination from normal mammary gland. In addition, the tissue immediately adjacent to each MIN lesion was primarily composed of fat, and the fat and water signals are separated by the fitting procedure used here; thus, any contamination from fat in the MIN ROI would not affect the measured T2* of water. Therefore, the short T2*’s reported here accurately reflect the properties of pure intraductal cancer. The results suggest that images of T2* in patients could be useful for detecting DCIS lesions. T2* images might also be useful in preclinical studies of mammary cancer development. Addition of T2* imaging to conventional breast MRI protocols may improve detection and diagnosis of DCIS/MIN. Although T2*s are much longer at the lower field strengths used for clinical imaging, T2* differences between DCIS and normal mammary gland may be detectable. In fact, previously published results from high spectral and spatial resolution MR imaging demonstrate intraductal cancer in images of the water resonance peak height.13
There is an apparent increase in the T2* of normal mammary glands of transgenic mice when fat suppression is applied. An intriguing explanation for this is that fat suppression pulses affect the broad component of the water resonance that underlies the fat signal—producing magnetization transfer (MT). This could change the relative contributions of the various populations of water molecules that make up the water resonance, and thus change the measured T2*, as well as other relaxation times. Fat suppression had no detectable effect on the measured T2* of water in MIN, tumor, lymph node, or muscle. The known magnetization transfer effects in fibroglandular breast tissue (20%) support the possibility of a change in T2* due to magnetization transfer.14 However, MT effects in pure intraductal cancer are not known, and therefore more research is needed to determine whether MT contributes to the observed changes in T2* of normal parenchyma with fat suppression. If there are differential effects of magnetization transfer on T2* in normal mammary gland vs intraductal cancer, it may be possible to make use of this effect as a diagnostic marker. T2* measurements combined with magnetization transfer may increase sensitivity and specificity for detection of early in situ cancer.
Although our work suggests that T2* could differentiate normal mammary gland and MIN, this result may not directly translate to clinical applications because the murine mammary gland is not a perfect model for human anatomy: human mammary glands develop within a fibroglandular matrix, surrounded by adipose tissues, while murine mammary glands may have a more modest fibrous component.
In summary, it is difficult to measure T2* for early in situ cancer in patients because these cancers are diffuse and have complicated anatomy. In the transgenic mouse model used here individual intraductal cancers can easily be identified and imaged at high resolution, and their T2*s can be accurately measured. T2* measurements could also be used in murine mammary models of breast cancer to follow cancer progression noninvasively. While these preliminary results are promising, they are based on an initial study of a small number of animals. To achieve better statistical certainty a larger cohort will be necessary. In addition, higher resolution multigradient echo images should be acquired to allow more accurate segmentation of individual mammary ducts. This study in a mouse model provides a rationale for similar studies of patients using T2* to diagnostic early breast cancer.
ACKNOWLEDGMENTS
The authors are grateful to the Segal Foundation, NIH Grant Nos. P50 CA125183-01 (SPORE), R33CA100996-02, R01CA133490-01A2, and The University of Chicago Cancer Center for financial support.
References
- Nelson H. D., Tyne K., Naik A., Bougatsos C., Chan B. K., and Humphrey L., “Screening for breast cancer: An update for the US Preventive Services Task Force,” Ann. Intern. Med. 151, 716–726 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson S., “Blood oxygenation level dependent (BOLD) imaging of tumors,” in New Techniques in Oncologic Imaging, Padhani A. R. and Choyke P., eds. (Taylor & Francis, Boca Raton, 2005), pp. 257–272. [Google Scholar]
- Foxley S., Fan X. B., Mustafi D., Haney C., Zamora M., Markiewicz E., Medved M., Wood A. M., and Karczmar G. S., “Sensitivity to tumor microvasculature without contrast agents in high spectral and spatial resolution MR images,” Magn. Reson. Med. 61, 291–298 (2009). 10.1002/mrm.v61:2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J. E., Shibata M. A., Yoshidome K., Liu M. L., Jorcyk C., Anver M. R., Wigginton J., Wiltrout R., Shibata E., Kaczmarczyk S., Wang W., Liu Z. Y., Calvo A., and Couldrey C., “The C3(1)/SV40 T-antigen transgenic mouse model of mammary cancer: Ductal epithelial cell targeting with multistage progression to carcinoma,” Oncogene 19, 1020–1027 (2000). 10.1038/sj.onc.1203280 [DOI] [PubMed] [Google Scholar]
- Jansen S. A., Conzen S. D., Fan X. B., Markiewicz E. J., Newstead G. M., and Karczmar G. S., “Magnetic resonance imaging of the natural history of in situ mammary neoplasia in transgenic mice: A pilot study,” Breast Cancer Res. 11, p. R65 (2009). 10.1186/bcr2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen S. A., Conzen S. D., Fan X., Krausz T., Zamora M., Foxley S., River J., Newstead G. M., and Karczmar G. S., “Detection of in situ mammary cancer in a transgenic mouse model: In vitro and in vivo MRI studies demonstrate histopathologic correlation,” Phys. Med. Biol. 53, 5481–5493 (2008). 10.1088/0031-9155/53/19/014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland A. E., Hendrick R. E., Jin H., Russ P. D., Barentsz J. O., and Holland R., “Correlation of high-resolution breast MR imaging with histopathology; Validation of a technique,” J. Magn. Reson. Imaging 11, 601–606 (2000). 10.1002/(ISSN)1522-2586 [DOI] [PubMed] [Google Scholar]
- Fan X. B., Abe H., Medved M., Foxley S., Arkani S., Zamora M. A., Olopade O. I., Newstead G. M., and Karczmar G. S., “Fat suppression with spectrally selective inversion vs. high spectral and spatial resolution MRI of breast lesions: Qualitative and quantitative comparisons,” J. Magn. Reson. Imaging 24, 1311–1315 (2006). 10.1002/(ISSN)1522-2586 [DOI] [PubMed] [Google Scholar]
- Foxley S., Fan X., Mustafi D., Yang C., Zamora M. A., Medved M., and Karczmar G. S., “Quantitative analysis of water proton spectral lineshape: A novel source of contrast in MRI,” Phys. Med. Biol. 53, 4509–4522 (2008). 10.1088/0031-9155/53/17/003 [DOI] [PubMed] [Google Scholar]
- Medved M., Du W. L., Zamora M. A., Fan X. B., Olopade O. I., MacEneaney P. M., Newstead G., and Karczmar G. S., “The effect of varying spectral resolution on the quality of high spectral and spatial resolution magnetic resonance images of the breast,” J. Magn. Reson. Imaging 18, 442–448 (2003). 10.1002/(ISSN)1522-2586 [DOI] [PubMed] [Google Scholar]
- Oikawa H., AlHallaq H. A., Lewis M. Z., River J. N., Kovar D. A., and Karczmar G. S., “Spectroscopic imaging of the water resonance with short repetition time to study tumor response to hyperoxia,” Magn. Reson. Med. 38, 27–32 (1997). 10.1002/(ISSN)1522-2594 [DOI] [PubMed] [Google Scholar]
- O’Regan D. P., Callaghan M. F., Wylezinska-Arridge M., Fitzpatrick J., Naoumova R. P., Hajnal J. V., and Schmitz S. A., “Liver fat content and T2*: Simultaneous measurement by using breath-hold multiecho MR imaging at 3.0 T—Feasibility,” Radiology 247, 550–557 (2008). 10.1148/radiol.2472070880 [DOI] [PubMed] [Google Scholar]
- Medved M., Newstead G. M., Abe H., Zamora M. A., Olopade O. I., and Karczmar G. S., “High spectral and spatial resolution MRI of breast lesions: Preliminary clinical experience,” Am. J. Roentgenol. 186, 30–37 (2006). 10.2214/AJR.04.1704 [DOI] [PubMed] [Google Scholar]
- Bonini R. H. M., Zeotti D., Saraiva L. A. L., Trad C. S., Filho J. M. S., Carrara H. H. A., de Andrade J. M., Santos A. C., and Muglia V. F., “Magnetization transfer ratio as a predictor of malignancy in breast lesions: Preliminary results,” Magn. Reson. Med. 59, 1030–1034 (2008). 10.1002/(ISSN)1522-2594 [DOI] [PubMed] [Google Scholar]