Figure 2.
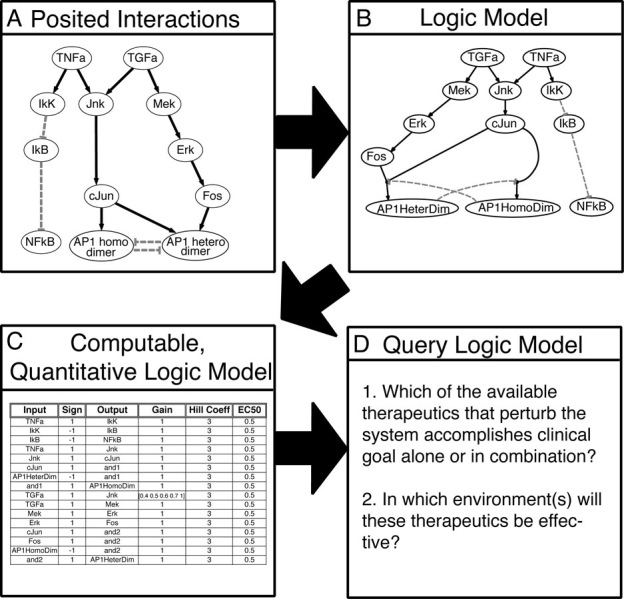
Converting posited interactions of intracellular signaling into a logic model. (A) The relationship between species in an intracellular signaling network is depicted graphically. Grey dashed blunted arrows indicate inhibitory interactions. (B) To convert the posited interactions in A into a logic model, we consider if the logic describing the relationship between input and output species should include an AND gate for species with more than one input, and find that AND gates are necessary for description of formation of AP1 homo- and heterodimers. AND gates are indicated by the input species linked to a small circle, which is further linked to the output species. (C) The logic model is recorded as a spreadsheet to be loaded into the Q2LM software. The first three columns specify which species interact as well as the logic of these relationships. The last three columns specify the parameters of the transfer functions of the interaction contained in that row. (D) Q2LM has been specifically designed to ask academically and industrially relevant questions.
