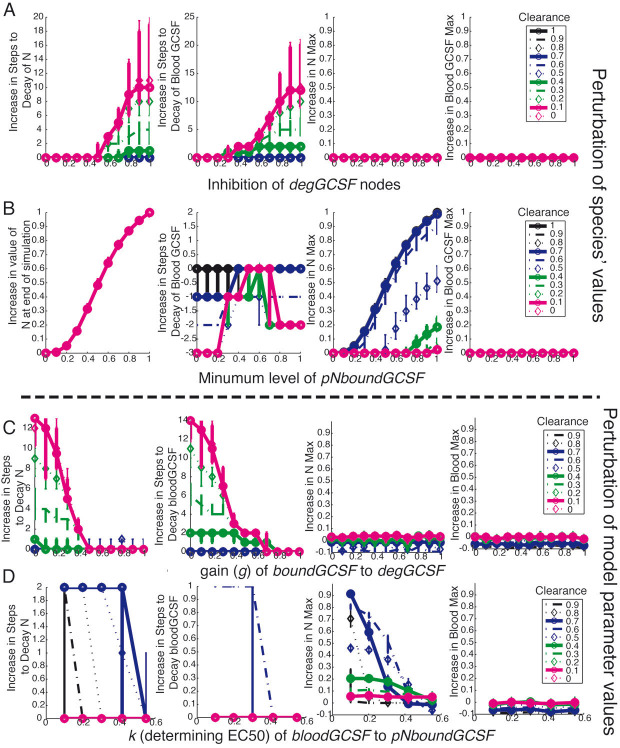
Figure 6.
Effect of perturbations to G-CSF pharmacokinetics on criteria. In all parts, perturbations to (A, B) species or (C, D) model parameters were made when the G-CSF logic model was simulated under non-limiting precursor neutrophils and dose conditions (i.e., pN = 1 and doseGCSF = 1) with multiple levels of clearance (0, 0.1, 0.2, etc.), with each color and line style corresponding to a different fixed value of the clearance species as shown in the legend in the rightmost panel for each part. Median effects are plotted, with error bars indicating the first and third quartile of predictions of 100 models. (A) The median effect of increasing inhibition of the pNdegGCSF and NdegGCSF nodes on each criteria. (B) The median effect of varying the minimal possible value of the pNboundGCSF node. Because the N species was not observed to decay in these simulation, the first panel is the increase in logic steady state value of N, instead of number of steps until decay. (C) The median effect of changing the gain of the transfer function relating pNboundGCSF to pNdegGCSF and NboundGCSF to NdegGCSF on each criteria. (D) The effect of changing the EC50 of the bloodGCSF to pNboundGCSF interaction.