Abstract
Age-related macular degeneration (AMD), affecting 30 to 50 million elder individuals worldwide, is a disease affecting the macular retina and choroid that can lead to irreversible central vision loss and blindness. Recent findings support a role for immunologic processes in AMD pathogenesis, including generation of inflammatory related molecules in the Bruch’s membrane, recruitment of macrophages, complement activation, microglial activation and accumulation in the macular lesions. Pro-inflammatory effects of chronic inflammation and oxidative stress can result in abnormal retinal pigment epithelium, photoreceptor atrophy and choroidal neovascularization. The associations of immunological and inflammatory genes, in particular the genes related to innate immunity with AMD support the involvement of various immunological pathways in the AMD pathogenesis. We review the literature on the involvements of inflammatory genes in AMD, highlight recent genetic discoveries, and discuss the potential application of such knowledge in the management of patients with AMD.
Keywords: age-related macular degeneration (AMD), complement factor, cytokine/chemokine, gene, innate immunity
Age-related macular degeneration (AMD) is a neurodegenerative disease of the central retina (the maculae) that represents the most common cause of irreversible visual impairment among people over age 50 in the world. Risk is multifactorial, relating to age and various environmental, dietary, and genetic factors. Heterogeneity of disease phenotype and onset late in life has complicated the disease mechanism. However, advances in genetic epidemiology and technology have recently transformed our understanding of heritable risk factors and provided insights into AMD pathophysiology. While AMD is not a classical inflammatory disease and inflammatory cells are reduced compared to other inflammatory tissues or are even neglected in these lesions, macrophages and even lymphocytes have been found in AMD lesions.1 There is a growing body of evidence suggesting an important role of inflammatory and immunological events in AMD pathogenesis.2–4
Clinical and histopathological hallmark of AMD is macular atrophy with drusen. Clinically, drusen are subretinal, discrete or soft yellowish lesions. Histopathologically, drusen are pancake-shaped, amorphous eosinophilic materials in Bruch’s membrane beneath retinal pigment epithelial cells (RPE),3 Biochemically, drusen contain lipids, proteins.5–7 and oxidation products from lipids and carbohydrates.6,8 These components from oxidative modifications in drusen and Bruch’s membrane can activate pattern recognition receptors that initiate inflammatory and immune responses.9 Large drusen and greater number of drusen in the maculae may develop geographic atrophy (‘dry’ AMD) and choroidal neovascularization CNV) or exudative/neovascular AMD (‘wet’ AMD). Drusen and the retinal lesions in advanced AMD can initiate and promote inflammatory responses, as well as innate and adaptive immune reactions, such as macrophage recruitment, microglial accumulation, complement activation, cytokines/chemokines releasing, and inflammatory oxidative stress.
Studies on twins and affected families have long suggested a heritable component to AMD risk.10,11 Genetic linkage studies found disease susceptibility loci on chromosomes 1q25–31 and 10q26.12 In the past 7 years, genome-wide and targeted genetic association studies have identified various polymorphisms important for AMD susceptibility.13–19 Notably, many of them are immune-related genes, including complement system, e.g. factor H (CFH), complement component 2 (C2), complement component 3 (C3), complement factor B (FB), and complement factor I (FI); toll-like receptor (TLR), e.g., TLR-3 and TLR-4; and chemokines, e.g., CX3CR1 and CCR3. However, controversies have been raised over the associations of TLR and AMD.20–22
I. AMD ASSOCIATED IMMUNOLOGICAL GENES IDENTIFIED BY LINKAGE AND FAMILY-BASED STUDIES
Data from most family studies of AMD have supported a strong genetic component to disease.23–25 Linkage studies have suggested several chromosomal regions that may harbor AMD susceptibility genes. Klein and colleagues were the first to map a susceptibility locus, ARMS1, to chromosome 1q25–q31 in a large pedigree with predominantly geographic atrophy AMD.24 This locus includes CFH.
Recent studies identified several candidate loci. Weeks et al reported potential loci on chromosomes 1q31, 9p13, 10q26, and 17q25.23 These results corroborate with Klein et al,24 in which linkage to chromosome 1q25–q31 shows an LOD score >1.5. Several of the following studies also confirmed the linkage on chromosome 1, mapping between 1q25–31.10–12 Schick et al. performed a genome-wide scan of 102 families from the Beaver Dam eye study and did not find a strong signal on chromosome 1, but reported 12q23–24, near D12S346, to have the strongest signal.26 Other potential regions included 5q12–13, 6q14–21, 15q11–14, 15q25–26. D5S2500, on chromosome 5, D12S1300 and PAH on chromosome 12, had all been previously reported.23 Seddon et al found significant linkage on chromosomes 2, 3, 6, and 8, in addition to 1, 10, 12, 16, 22, X.11 Mejewski et al studied 70 large families and found evidence for linkage in five regions with scores exceeding LOD = 2 − 1q31, 3p13, 4q32, 9q33 and 10q26.10 Several of these regions are close to the chromosomal position of candidate immune genes associated with AMD (Table 1). Iyengar et al performed a genome wide scan on 34 extended pedigrees and had their strongest signal at 15q14, and observed 13 other regions on 11 chromosomes,27 many of which were also consistent with previous reports. Abecaiss et al performed a genome wide scan of 412 affected relative pairs and found linkage in 5 regions: 1q41, 2p25, 5p13–14, 9q31, 22q12.28
Table 1.
Candidate immune genes associated with AMD.
| Gene symbol (name) | Function | Chromosomal Position |
Variant | Odds Ratios (OR) |
References |
|---|---|---|---|---|---|
| CFH (Complement factor H) | Inhibitor of alternative complement pathway | 1q32 | rs1061170 | OR hm = 6.32 | 14, 16, 17 |
| C2/CFB (Complement component 2/Complement factor B) | Regulation of complement activation | 6p21 | rs9332739 (C2) rs4151667 (CFB) rs641153 (CFB) |
OR ht = 0.32–0.40 | 42, 44 |
| CFHR1/CFHR3 (Complement factor H-related 1, 3) | Unknown, possible overlapping function with CFH | 1q31-q32 | 84K bp deletion | OR hm = 0.29 | 38, 83 |
| C3 (Complement component 3) | Innate immunity (alternative complement pathway activator, classical pathway component) | 19p13 | rs2230199, rs1047286 |
OR hm = 1.93–3.91 | 45 |
| C5 | Innate immunity (alternative complement pathway component) | 9q33–q34 | rs17611, rs7026551, rs7037673 |
OR hm = 0.66O R hm = 1.50O R = 0.51 |
116 |
| HLA (Human leukocyte antigen) class I and II | Regulation of immune response | 6p21.3 | Cw*0701 DQB1*0303 |
OR hm = 1.85O R hm = 2.67 |
117 |
| CX3CR1 (Chemokine [C-X3-C motif] receptor 1) | Inflammation (chemokine receptor) | 3p21 | rs3732378 | OR hm = 1.98–2.70 | 47, 48 |
| TLR3 (Toll-like receptor 3) | Innate immunity (targets + viral dsRNA) | 4q35 | rs3775291 | OR hm = 0.44–0.61 | 19 |
| TLR4 (Toll-like receptor 4) | Innate immunity (bacterial endotoxin receptor) | 9q32–q33 | rs4986790 | Conflicting results:OR ht = 2.65; no association |
118, 119 |
OR hm homozygous odds ratio for risk allele; OR ht heterozygous odds ratio for risk allele
Both 1q15–31 and 10q26 have showed the most consistency and have since been validated.10,11,24,29 Region 1q15–31 has shown implication in at least 5 studies and 10q26 also been implicated in several studies. Since these studies, CFH at 1q32 has been shown to be associated with AMD. In addition, HTRA1 and ARMS2/LOC387715, located at 10q26, show strong association with AMD (Table 1).
Several other linkage studies have searched for new candidate genes. With evidence suggestive of another AMD locus on chromosome 16p12, one group studied five genes within this locus – CACNG3, HS3ST4, IL4R, Q7Z6F8, ITGAM. The strongest evidence for linkage was within CACNG3. After adjusting for known AMD risk factors, rs2283550 remained the most strongly associated SNP.30 One linkage study reported SKIV2L and MYRIP as protective factors for AMD.31 In addition, LIPC and TIMP3 were suggested to be associated with AMD.32 A susceptibility locus was identified near TIMP3, which is an extracellular matrix metalloproteinase that has been implicated in early onset maculopathy.33 The association of loci within LIPC and AMD has suggested a connection between HDL metabolism and AMD pathogenesis.
These studies have shown that the use of families with significant history of AMD has also been quite effective in the discovery of genes associated with AMD. However, evaluation of the genetic basis of AMD through family studies has its challenges. Many individuals in a family are older and may have died, or they suffer from a multitude of other diseases that complicate results. In addition, many members may be younger and have not developed signs of macular degeneration. Therefore, several generations of informative individuals are not easily available.
II. AMD ASSOCIATED IMMUNOLOGICAL GENES IDENTIFIED BY GENOME-WIDE AND CANDIDATE GENE ASSOCIATION STUDIES
Genome-wide association study (GWAS) studies have revealed CFH to have a strong association to AMD.13,15,34 Studies have shown an increase risk of AMD in individuals with a T>C substitution in exon 9 of CFH representing a tyrosine-histidine change at amino acid 402 (Y402H). This polymorphism has been located to a region of CFH that binds heparin and C-reactive protein.13,35 Another study found association of 8 common SNPs in the CFH gene, with two missense mutations exhibiting high significance to AMD: 162V and also the Y402H variant.15 The strongest association was found for the synonymous A473A (rs2274700:G>A) variant in exon 10 with an odds ratio of 3.42, 95% CI 2.27–5.15. These SNPs have been confirmed in numerous studies.16,36–38
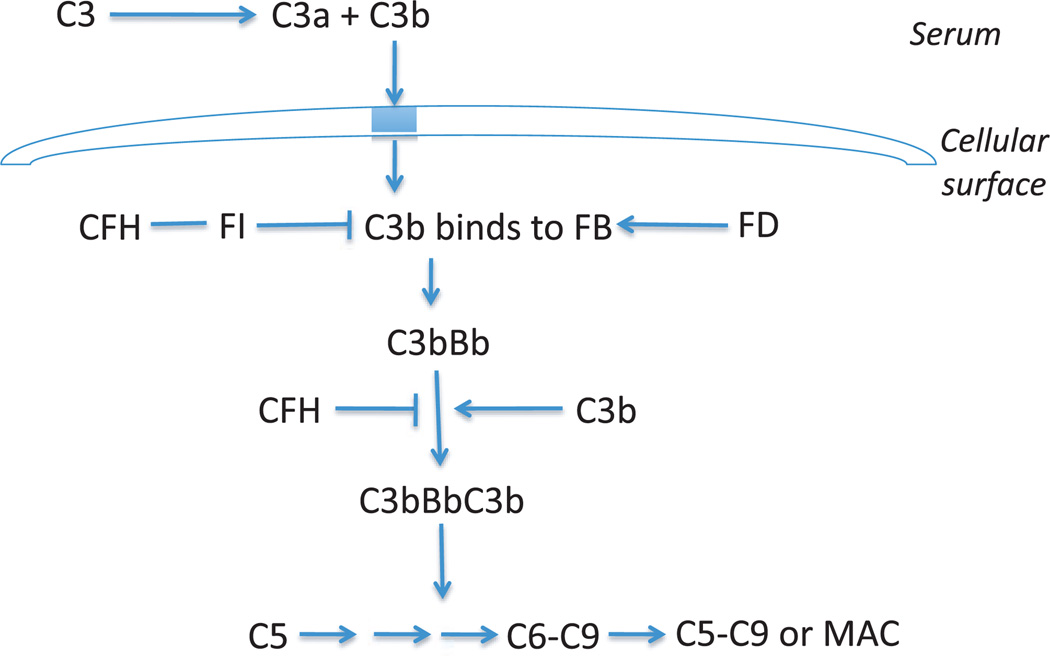
At least 9 of the loci encode proteins of the immune-related pathways found to be associated with AMD–CFH, FB/C2, C3, C5, human leukocyte antigen (HLA) locus, CX3CR1, CCR3, and TLR3, TLR4 (Table 1). Therefore, the immune system is proposed to play a central role in the pathogenesis of AMD. CFH is an important regulator of the complement system. The activation of the alternative complement cascade is initiated by the formation of the C3Bb complex, also known as C3 convertase (Figure 1). Formation of this complex leads to the development of the terminal complex and amplification of the immune response. The dissociation of this C3Bb complex is accelerated by CFH. Therefore, alterations in the structure of CFH can lead to abnormal or increased immune activity. Studies have implicated local inflammation and activation of the complement cascade in the formation of drusen, a hallmark of early AMD.39 Similar inflammatory mechanisms have been reported in relation to formation of extracellular plaques in Alzheimer’s disease and deposits in atherosclerosis.39,40
Figure 1.
Alternative Pathway C3, complement component 3; CFH, complement factor H; FI, factor I; FB, factor B; FD, factor D; C5, complement component 5; C6, component component 6; C9, complement component 9; MAC, membrane attack complex.
Both components of the complement system have been found in drusen of AMD patients and have been shown to contribute to vascular endothelial growth factor (VEGF) expression, a signal for angiogenesis.41 Recent GWAS studies have further confirmed the association of C3 with AMD.20 FB and C2 genes were associated with decreased susceptibility to AMD.42–44 FB and C2 function as activators of the alternative and classic complement pathways, respectively. If their enzymatic activity is reduced due to alterations in protein structure, then these alternations may provide a lower risk of chronic complement activation and therefore, a decreased risk of AMD.
Different variants within C3 and FB/C2 have been identified to be associated with AMD.42–45 The R32Q variant of FB showed protective from AMD in a family based study. Three SNPs in C2 and FB were strongly associated with decreased risk of AMD in a case-control study.44 The minor alleles at C2 rs547154 and FB R32Q were present in 4% of cases versus 10% of the controls.44 Complement C3 S/F (Arg80Gly) also showed strong evidence of an association with AMD.45 These studies further support the role of the complement pathway in AMD development. However, functional studies are needed to confirm the protective effects of these genes on AMD development.
Other immune-related genes associated with AMD are CX3CR1, likely related to the recruitment of macrophage and microglia. Three independent studies showed a mild association of CX3CR1 variants with AMD.46–48 However, Ryu et al did not find this association in a recent GWAS study.20 On another recent large population of 1,093 neovascular AMD patients compared to 396 controls in France, no association was reported.49 Takeda et al. reported that CCR3, an eosinophil/mast cell chemokines receptor, was highly expressed in human CNV membrane, but not in normal vessels. These authors suggested that CCR3 could be an early marker of and potential therapeutic target for CNV.50 However, Li et al demonstrated no inhibitory effect of CCR3 antibody on CNV using the matrigel induced model in mice and rats.21
III. GENE–ENVIRONMENT AND GENE–GENE INTERAC TIONS
AMD is a complex multifactor disease involving both genetic and environmental risk factors. Studies have shown that gene-gene and gene-environment interactions play a role in disease risk. These interactions have been studied in order to provide further information about the contribution of risk factors to the development of AMD. Of the environmental risk factors, age and smoking have been the most consistent.32 Body mass index (BMI) has shown a marginal association.51 It has been accepted for many years that the immune system undergoes changes resulting in loss of immune function with age. Smoking is known to weaken the immune system and has been confirmed as a risk factor for AMD. Patients have a 1.8-fold higher chance of AMD if they ever smoked compared to those who never smoked.51 However, it still remains unclear as to the extent of their effect on AMD development. Weeks et al hypothesized that the effects of smoking on the risk of AMD is accentuated by a gene in region 10q26.12 Since then, the joint effects of genetic and environmental factors are implicated to have a better prediction for advanced AMD.52–58
Schmidt et al found that smoking strongly modifies the association of LOC387715 and AMD and estimated that CFH, LOC387715 and cigarette smoking together explain 61% of the population-attributable risk of AMD.53 Seitsonen et al found that smoking exerted an extra risk for AMD,52 but seemed to only in connection with other factors such as sex and C3 genotype. Nakanishi et al studies suggested interactions between CFH Y402H and cigarette smoking.57 Shaumberg et al also suggested interplay of genetic and environmental risk factors stating that patients homozygous for the risk alleles of CFH Y402H and LOC387715 A69S had a 50-fold increase in the risk of AMD and cigarette smoking and obesity multiplied the risk as well.54
As for gene-gene interactions, there have been differing conclusions. Chen et al found weak interactions between CFH rs1061170 and HTRA1/LOC387715 rs10490924 as well as between CFH rs2274700 and HTRA1/LOC387715 rs10490924.51 This result was also similar in a study in Finland, which showed a possible interaction between CFH and LOC387714.52 However, other studies have not found this interaction.18,53,56,59,60 The intricate interplay of genetics and environmental factors will need to be further studied in order to better understand people’s risk for AMD.
IV. DISTINCT AMD LOCI IN ETHNIC GROUPS
Several studies have looked at specific populations to determine if the susceptibility genes could be generalized to different locations and ethnicities or if other genes played a more significant role. For example, in a French population PLEKHA1 (rs4146894), HTRA1 (rs11200638) and ARMS2/LOC387715 (rs10490924) were found to be independently and strongly associated with exudative AMD.61 A study of an Indian population validated the association of risk alleles within ARMS2/LOC387715 (rs10490924) and HTRA1 SNPs (A allele: rs11200638 and C allele: rs2672598), along with risk estimates among Indian patients with AMD.62 A study found statistically significant associations of three SNPs of ARMS2/HTRA1 region: rs3793912, rs10490924, rs11200638 and AMD in Japanese population.63 Chu et al found CFH and HTRA1 polymorphisms to be potentially additive contributors to exudative AMD in the Han Chinese population.64 Another study focusing on a separate group of Singaporean Chinese indicated that the TLR3 rs3775291 is not associated with CNV and polypoidal choridal vasculopathy (PCV), a condition similar to but distinct from AMD.65
V. COPY NUMBER VARIATION OF IMMUNE RELEVANT GENES AND AMD
Although numerous SNPs have been discovered to be highly associated with AMD, they do not account for the entire genetic component of the disease. Interindividual variation in human genome exists not only in the sequence level but also in other structures. Deletions and duplications can result in alterations in the copy number of affected segments of the human genome.66 Copy number variation usually refers to large repeated regions of more than several hundred nucleotides and distinguishes from microsatellites and minisatellites.67 Copy number variation deviated from normal two copies could be dialleletic or multialleletic. However, due to the lack of accurate technique, copy number variation genotypes are often reported as “gain” or “loss” relative to dialleletic type. Population-based studies have identified thousands of copy number variation loci throughout the human genome.67,68 The proposed mechanisms on copy number variation impact include gene dosage, gene interruption, gene fusion, and positional effects.69 Copy number variation of dosage-sensitive genes may cause or predispose to a variety of human diseases. The association of copy number variation have been reported with disease susceptibility, among most of which were immune related disorders including glomerulonephritis,70 psoriasis,71,72 systemic lupus erythematosus,71,73–75 and HIV.76
While copy number variation contributes to the susceptibility of various human diseases, only a limited number of studies on its role in AMD pathogenesis were reported. GWAS failed to identify any copy number variation loci associated statistically significant with AMD; however, those studies spotted several rare copy number variations close to or in known AMD susceptible genes such as EFEMP1.77–80 Moreover, direct sequencing of the targeting loci and multiplex ligation-dependent probe amplification was able to find common deletions in CFH related genes 1 and 3 (CFHR1 and CFHR3), which are protective against AMD.38,81–83 Individuals who carry the homozygous deletion had no detectable CFHR1 protein in their serum, indicating the functional importance of the structural variation, and showed no evidence of AMD. Even though the boundary of the deletion has not been precisely determined yet, it lies within the regulators of complement activation locus on chromosome 1q32. Combination of two large cohorts in Caucasian recorded a 5.7% of frequency of the homozygous CFHR1 deletion in the non-AMD population with an odds ratio of 0.191 to AMD.81 The deletion of CFHR1 allele also displays considerable variation between racial/ethnic groups. Homozygotes are most common in African Americans (16%), less common in Hispanics (6.8%). The district distribution of this protective factor between ethnic groups is in agreement with the low frequency of late-onset AMD in African Americans, compared to the Caucasians.84
When there is no clue to the potential contributor of copy number variation of any genes in AMD pathogenesis, it is logical to place the precedence for exploring copy number variations of genes that have been identified to be SNP-AMD-associated. Similar approach has been demonstrated to be successful in the study of α-synuclein gene in Parkinson’s disease.85,86 There is also evidence for linkage disequilibrium between copy number variations and SNPs in human genome.87 We performed quantitative copy number genotyping for several AMD genes in neovascular AMD patients and elderly controls.88 Among 6 AMD relevant genes, CCR3, CFH, CX3CR1, VEGF, ERCC6, and HTRA1, 4 of them function as immune molecules. By analyzing 131 persons with neovascular AMD and 103 elderly persons without AMD, we found that copy number variations of above genes existed in both AMD and control populations. As estimated by a maximum likelihood algorithm, based on the probability density distribution across all samples, the copy number variation = 2 was the predominant copy number genotype of all copy number variations of the tested genes. Novel copy number variations were found within CCR3 and CX3CR1. Even though the copy number variations of the tested genes did not differ between the AMD and control after strict statistical justification, there were trends in the unadjusted data suggesting that CX3CR1 might be a gene of interest in terms of copy number variation. The rates of copy number = 3+ carriers for CX3CR1 were 25.0% in AMD and 14.6% in controls (OR = 0.52, 95% CI: 0.27–1.01, p = 0.05), indicating a protective role of the extra copy of CX3CR1 for AMD.88 The result is in agreement with previous reports that two loss-of-function SNPs in CX3CR1 are associated with moderately elevated AMD risk.46–48 In addition, mice with Cx3cr1 or Cx3cr1/Ccl2 being knocked out develop spontaneously AMD-like pathological features in retina.46,89,90
VI. PHARMACOGENETICS OF IMMUNE RELEVANT GENES AND AMD
In the past decade, some new treatment and prevention options have been introduced in an attempt to minimize the AMD-induced damages. A high-dose of an orally administered combination of the vitamin C, vitamin E and beta-carotene, in addition to copper and zinc, is a widely accepted preventive approach.91 Thermal laser photocoagulation and verteporfin photodynamic therapy (PDT) are also the extensively used options.92 Currently, the most widely utilized therapy for neovascular AMD is intravitreal administration of anti-VEGF antibodies. However, patients do not equally respond to these treatments. Some suggested that the AMD-susceptibility SNPs or even SNPs in genes functioning in the therapy-related pathways are responsible for the differentiated responses.93–100
A study on the interaction between the CFH Y402H/ARMS2 A69S variants and supplementation with antioxidants plus zinc found that in individuals with homozygous CFH non-risk variant (402YY), 34% in the placebo group progressed to advanced AMD, compared with 11% in the antioxidants plus zinc-treated group: a reduction of approximately 68%. Of those individuals with the homozygous CFH risk variant (402HH), 44% in the placebo group progressed to advanced AMD, compared with 39% in the anti-oxidants plus zinc-treated group: a reduction of only 11%, indicating that CFH non-risk variant carriers have a better response to the intervention than risk variant carriers. In addition, a similar interaction was observed in the groups taking zinc alone versus those taking no zinc. Of those 402YY carriers, 36% in the no zinc group progressed to advanced AMD, compared with 14% in the zinc-treated group; a reduction of 61%. Of those 402HH carriers, 47% in the no zinc group progressed to advanced AMD, compared with 42% in the zinc-treated group; a reduction of only 11%.98 The study did not find interaction between the ARMS2 A69S variant and the AREDS treatment, indicating that inflammatory factors are not only involved in the AMD susceptibility but also in individual’s response to nutrient supplements. Specific biological mechanisms have not yet explained the interaction and that CFH variants had an altered binding affinity to zinc.101
Another study found that individual’s carrying non-risk CFH 402YY had worse outcomes than that of risk CFH 402 carriers after PDT.102,103 Among the 27 patients eligible to PDT, the average visual loss following PDT was 70 letters in 402YY carriers (n = 2), of 3.5 letters in 402YH carriers (n = 12) and 12 ETDRS letters in 402HH carriers (n = 13).102 Two independent studies exhibited similar results in terms of average post-PDT visual acuity and proportion of responders.99,104,105 Interestingly, patients with unfavorable genetic makeup for AMD may have better outcome after PDT. One explanation is that patients with the non-risk CFH variant may develop CNV by other non-inflammation mechanisms rather than complement factors, and this makes them less responsive to PDT.105 Another AMD-susceptibility SNP, ARMS2 A69S, did not show any modifying effect on the PDT therapy.105
VEGF is recognized as a key mediator in the CNV formation during AMD development. Poor visual acuity after anti-VEGF therapy was found in AMD patients carrying risk CFH 402HH compared to the other CFH402 genotypes combined after giving bevacizumab. However, there was no association between the response to bevacizumab and ARMS2 A69S genotypes. 99 A similar study on exudative AMD treated with ranibizumab found that CFH 402HH carriers had a 37% higher chance of requiring additional ranibizumab injections because of recurrence.96 Even though different outcomes were adopted in evaluation, similar relationships were identified in a prospective study, which observed a significantly worse outcome for distance and reading visual acuity in the CFH 402HH genotype group.100 However, a recent study reported a trend of more favorable visual acuity outcomes after 6 months of intravitreal ranibizumab therapy in patients carrying AMD risk-variants of CFH, VEGF and HTRA1,106 which is controversial to previous studies.96,99,100 No explanation for this discrepancy was offered in the study.106
Instead of only focusing on the SNPs with AMD susceptibility and the outcomes of anti-VEGF therapy, we recently conducted a study to determine pharmacogenomic mechanism of some competent immunological molecules among others, including CFH, HTRA1, IL-17, IL-23R, CYP3A, LEP and VEGFA. Initial data indicated a trend of association of an IL-23R SNP in the response to anti-VEGF therapy. This result echoes a novel finding of the involvement of IL-17 pathway in AMD pathogenesis.107
While the concept of AMD pharmacogenomics is promising, the progress in this field is lagging. The available literatures on this field are based on studies with small sample size, heterogeneous study participants and non-standardized therapy protocol. The definitions for therapy response are ambiguous and long-term follow-up is lacking. The study in retrospective design is another problem. Moreover, when majority focusing on the interaction of AMD-susceptibility SNPs with therapeutic responses, it should be understood that possible genetic components on the therapy responses might not be necessarily associated with the disease susceptibility itself. Some studies selected genes functioning in the activation of therapy-induced pathways to test the possible modifying effect. As cited before, SNPs in coagulation balance genes (factor V, prothrombin) may affect the efficacy of PDT.93,95 Similar approaches can be used by carefully selecting functional variations in the inflammation related genes and test their role in therapy responses.
VII. AMD AND IMMUNE REGULATION BY EPIGENETICS AND MicroRNAS
In general, epigenetics refers to the study of heritable changes in gene expression caused by mechanisms other than changes in DNA sequence. Examples of such changes are DNA methylation and histone acetylation, both of which suppress gene expression. The influence of epigenetic changes in common complex diseases has been extensively reported.108 Collective evidence indicates modulations of environmental pressure on gene expression in the immune system through epigenetic mechanisms.109 However, thus far, there is no peer-reviewed full article published on epigenetics and AMD. Two presentations in ARVO 2011 demonstrated an altered epigenetic profile of immune genes in ocular tissue from AMD patients by large scale epigenetic array. One of the studies applied a twin-based design by recruiting two pairs of monozygotic twins with discordant AMD pathology. Among 2.1 million gene promoters, the study identified ~1000 candidate genes within which promoters differential DNA methylation patterns were found. Among those 1000 genes, 256 genes were associated with hypomethylated CpG sites, while 744 genes with hypermethylated CpG sites were only in twins with AMD. One exemplar locus of those differentially methylated loci is within CCL22 locus. Hypermethylated CpG sites were only found in control but not AMD twins, indicating a potential role of CCL22 in AMD.110
MicroRNAs (miRNAs) are non-protein-coding RNAs and functions as post-transcriptional regulators that bind to complementary sequences on target mRNAs. miRNA usually functions as translational repressor and gene silencing. The human genome may encode over 1000 miRNAs and target about 60% of mammalian genes.110 miRNAs appear to regulate the innate and adoptive immune systems. Aberrant miRNA expression can contribute to pathological conditions involving the immune system.111 A preliminary study described immune gene regulation by miRNA in retinal tissue.112
A study reported in ARVO 2011 found district patterns of miRNA expression in peripheral blood monocytes of geographic atrophy AMD patients. The study suggests the involvement of systemic immune responses in the pathogenesis and progression of geographic atrophy AMD.
Several animal studies reported the involvement of miRNAs in AMD relevant pathological features. Accumulation of miRNA in retina due to disruption of DICER1 can cause retinal lesion mimicking geographic atrophy AMD.113,114 miR-23~27~24 gene clusters can regulate angiogenesis and choroidal neovascularization, since miRNAs encoded by the miR-23~27~24 gene clusters are enriched in endothelial cells and highly vascularized tissues.115
In summary, genetic studies have mapped genes in the complement pathway that are involved in the regulation of innate immunity with AMD susceptibility and pharmacogenesis. Majority of the association in complement genes have replicated in diverse populations worldwide. Gene-gene and gene-environment interactions are also found as significant covariates in AMD pathology. Studies of copy number variation, epigenetics and miRNA may provide further evidence on the underlying molecular genetic mechanisms in AMD.
Acknowledgement
The NEI Intramural Research program provided funding.
Footnotes
Declaration of interest: The authors report no conflicts of interest
REFERENCES
- 1.Penfold PL, Killingsworth MC, Sarks SH. Senile macular degeneration: the involvement of immunocompetent cells. Graefes Arch Clin Exp Ophthalmol. 1985;223:69–76. doi: 10.1007/BF02150948. [DOI] [PubMed] [Google Scholar]
- 2.Anderson DH, Radeke MJ, Gallo NB, et al. The pivotal role of the complement system in aging and age-related macular degeneration: hypothesis re-visited. Prog Retin Eye Res. 2010;29:95–112. doi: 10.1016/j.preteyeres.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ding X, Patel M, Chan CC. Molecular pathology of age-related macular degeneration. Prog Retin Eye Res. 2008;28:1–18. doi: 10.1016/j.preteyeres.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel M, Chan CC. Immunopathological aspects of age-related macular degeneration. Semin Immunopathol. 2008;30:97–110. doi: 10.1007/s00281-008-0112-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crabb JW, Miyagi M, Gu X, et al. Drusen proteome analysis: an approach to the etiology of age-related macular degeneration. Proc Natl Acad Sci U S A. 2002;99:14682–14687. doi: 10.1073/pnas.222551899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L, Clark ME, Crossman DK, et al. Abundant lipid and protein components of drusen. PLoS One. 2010;5:e10329. doi: 10.1371/journal.pone.0010329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hollyfield JG, Salomon RG, Crabb JW. Proteomic approaches to understanding age-related macular degeneration. Adv Exp Med Biol. 2003;533:83–89. doi: 10.1007/978-1-4615-0067-4_11. [DOI] [PubMed] [Google Scholar]
- 8.Mullins RF, Hageman GS. Human ocular drusen possess novel core domains with a distinct carbohydrate composition. J Histochem Cytochem. 1999;47:1533–1540. doi: 10.1177/002215549904701205. [DOI] [PubMed] [Google Scholar]
- 9.West XZ, Malinin NL, Merkulova AA, et al. Oxidative stress induces angiogenesis by activating TLR2 with novel endogenous ligands. Nature. 2010;467:972–976. doi: 10.1038/nature09421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Majewski J, Schultz DW, Weleber RG, et al. Age-Related Macular Degeneration-a Genome Scan in Extended Families. Am J Hum Genet. 2003;73:540–550. doi: 10.1086/377701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seddon JM, Santangelo SL, Book K, Chong S, Cote J. A genomewide scan for age-related macular degeneration provides evidence for linkage to several chromosomal regions. Am J Hum Genet. 2003;73:780–790. doi: 10.1086/378505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weeks DE, Conley YP, Tsai HJ, et al. Age-related maculopathy: a genomewide scan with continued evidence of susceptibility loci within the 1q31, 10q26, and 17q25 regions. Am J Hum Genet. 2004;75:174–189. doi: 10.1086/422476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dewan A, Liu M, Hartman S, et al. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science. 2006;314:989–992. doi: 10.1126/science.1133807. [DOI] [PubMed] [Google Scholar]
- 14.Edwards AO, Ritter IR, Abel KJ, et al. Complement Factor H Polymorphism and Age-Related Macular Degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 15.Hageman GS, Anderson DH, Johnson LV, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci USA. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haines JL, Hauser MA, Schmidt S, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 17.Klein RJ, Zeiss C, Chew EY, et al. Complement Factor H Polymorphism in Age-Related Macular Degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rivera A, Fisher SA, Fritsche LG, et al. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum Mol Genet. 2005;14:3227–3236. doi: 10.1093/hmg/ddi353. [DOI] [PubMed] [Google Scholar]
- 19.Yang Z, Stratton C, Francis PJ, et al. Toll-like receptor 3 and geographic atrophy in age-related macular degeneration. N Engl J Med. 2008;359:1456–1463. doi: 10.1056/NEJMoa0802437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryu E, Fridley BL, Tosakulwong N, Bailey KR, Edwards AO. Genome-wide association analyses of genetic, phenotypic, and environmental risks in the age-related eye disease study. Mol Vis. 2010;16:2811–2821. [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Huang D, Xia X, et al. CCR3 and choroidal neovascularization. PLoS One. 2011;6:e17106. doi: 10.1371/journal.pone.0017106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho Y, Wang JJ, Chew EY, et al. Toll-like receptor polymorphisms and age-related macular degeneration: replication in three case-control samples. Invest Ophthalmol Vis Sci. 2009 doi: 10.1167/iovs.09-3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weeks DE, Conley YP, Mah TS, et al. A full genome scan for age-related maculopathy. Hum Mol Genet. 2000;9:1329–1349. doi: 10.1093/hmg/9.9.1329. [DOI] [PubMed] [Google Scholar]
- 24.Klein ML, Schultz DW, Edwards A, et al. Age-related macular degeneration. Clinical features in a large family and linkage to chromosome 1q. Arch Ophthalmol. 1998;116:1082–1088. doi: 10.1001/archopht.116.8.1082. [DOI] [PubMed] [Google Scholar]
- 25.Dosso AA, Bovet J. Monozygotic twin brothers with age-related macular degeneration. Ophthalmologica. 1992;205:24–28. doi: 10.1159/000310307. [DOI] [PubMed] [Google Scholar]
- 26.Schick JH, Iyengar SK, Klein BE, et al. A whole-genome screen of a quantitative trait of age-related maculopathy in sibships from the Beaver Dam Eye Study. Am J Hum Genet. 2003;72:1412–1424. doi: 10.1086/375500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iyengar SK, Song D, Klein BE, et al. Dissection of genomewide-scan data in extended families reveals a major locus and oligogenic susceptibility for age-related macular degeneration. Am J Hum Genet. 2004;74:20–39. doi: 10.1086/380912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abecasis GR, Yashar BM, Zhao Y, et al. Age-related macular degeneration: a high-resolution genome scan for susceptibility loci in a population enriched for late-stage disease. Am J Hum Genet. 2004;74:482–494. doi: 10.1086/382786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weeks DE, Conley YP, Tsai HJ, et al. Age-related maculopathy: an expanded genome-wide scan with evidence of susceptibility loci within the 1q31 and 17q25 regions. Am J Ophthalmol. 2001;132:682–692. doi: 10.1016/s0002-9394(01)01214-4. [DOI] [PubMed] [Google Scholar]
- 30.Spencer KL, Olson LM, Schnetz-Boutaud N, et al. Dissection of chromosome 16p12 linkage peak suggests a possible role for CACNG3 variants in age-related macular degeneration susceptibility. Invest Ophthalmol Vis Sci. 2011;52:1748–1754. doi: 10.1167/iovs.09-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kopplin LJ, Igo RP, Jr, Wang Y, et al. Genome-wide association identifies SKIV2L and MYRIP as protective factors for age-related macular degeneration. Genes Immun. 2010;11:609–621. doi: 10.1038/gene.2010.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen W, Stambolian D, Edwards AO, et al. Genetic variants near TIMP3 and high-density lipoprotein-associated loci influence susceptibility to age-related macular degeneration. Proc Natl Acad Sci U S A. 2010;107:7401–7406. doi: 10.1073/pnas.0912702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weber BH, Vogt G, Pruett RC, Stohr H, Felbor U. Mutations in the tissue inhibitor of metalloproteinases-3 (TIMP3) in patients with Sorsby’s fundus dystrophy. Nat Genet. 1994;8:352–356. doi: 10.1038/ng1294-352. [DOI] [PubMed] [Google Scholar]
- 34.Zareparsi S, Branham KE, Li M, et al. Strong Association of the Y402H Variant in Complement Factor H at 1q32 with Susceptibility to Age-Related Macular Degeneration. Am J Hum Genet. 2005;77:149–153. doi: 10.1086/431426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu Z, Brodsky B, Inouye M. Dissecting a bacterial collagen domain from Streptococcus pyogenes: sequence and length-dependent variations in triple helix stability and folding. J Biol Chem. 2011;286:18960–18968. doi: 10.1074/jbc.M110.217422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Francis PJ, Schultz DW, Hamon S, et al. Haplotypes in the complement factor H (CFH) gene: associations with drusen and advanced age-related macular degeneration. PLoS One. 2007;2:e1197. doi: 10.1371/journal.pone.0001197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li M, Atmaca-Sonmez P, Othman M, et al. CFH haplotypes without the Y402H coding variant show strong association with susceptibility to age-related macular degeneration. Nat Genet. 2006;38:1049–1054. doi: 10.1038/ng1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hughes AE, Orr N, Esfandiary H, et al. A common CFH haplotype, with deletion of CFHR1 and CFHR3, is associated with lower risk of age-related macular degeneration. Nat Genet. 2006;38:1173–1177. doi: 10.1038/ng1890. [DOI] [PubMed] [Google Scholar]
- 39.Mullins RF, Russell SR, Anderson DH, Hageman GS. Drusen associated with aging and age-related macular degeneration contain proteins common to extracellular deposits associated with atherosclerosis, elastosis, amyloidosis, and dense deposit disease. FASEB J. 2000;14:835–846. [PubMed] [Google Scholar]
- 40.Anderson DH, Talaga KC, Rivest AJ, et al. Characterization of beta amyloid assemblies in drusen: the deposits associated with aging and age-related macular degeneration. Exp Eye Res. 2004;78:243–256. doi: 10.1016/j.exer.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 41.Nozaki M, Raisler BJ, Sakurai E, et al. Drusen complement components C3a and C5a promote choroidal neovascularization. Proc Natl Acad Sci USA. 2006;103:2328–2333. doi: 10.1073/pnas.0408835103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gold B, Merriam JE, Zernant J, et al. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat Genet. 2006;38:458–462. doi: 10.1038/ng1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maller J, George S, Purcell S, et al. Common variation in three genes, including a noncoding variant in CFH, strongly influences risk of age-related macular degeneration. Nat Genet. 2006;38:1055–1059. doi: 10.1038/ng1873. [DOI] [PubMed] [Google Scholar]
- 44.Spencer KL, Hauser MA, Olson LM, et al. Protective effect of complement factor B and complement component 2 variants in age-related macular degeneration. Hum Mol Genet. 2007;16:1986–1992. doi: 10.1093/hmg/ddm146. [DOI] [PubMed] [Google Scholar]
- 45.Yates JR, Sepp T, Matharu BK, et al. Complement C3 variant and the risk of age-related macular degeneration. N Engl J Med. 2007;357:553–561. doi: 10.1056/NEJMoa072618. [DOI] [PubMed] [Google Scholar]
- 46.Combadiere C, Feumi C, Raoul W, et al. CX3CR1-dependent subretinal microglia cell accumulation is associated with cardinal features of age-related macular degeneration. J Clin Invest. 2007;117:2920–2928. doi: 10.1172/JCI31692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tuo J, Smith B, Bojanowski CM, et al. The involvement of sequence variation and expression of CX3CR1 in the pathogenesis of age-related macular degeneration. FASEB J. 2004;18:1297–1299. doi: 10.1096/fj.04-1862fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang X, Hu J, Zhang J, Guan H. Polymorphisms in CFH, HTRA1 and CX3CR1 confer risk to exudative age-related macular degeneration in Han Chinese. Br J Ophthalmol. 2010;94:1211–1214. doi: 10.1136/bjo.2009.165811. [DOI] [PubMed] [Google Scholar]
- 49.Zerbib J, Puche N, Richard F, et al. No association between the T280M polymorphism of the CX3CR1 gene and exudative AMD. Exp Eye Res. doi: 10.1016/j.exer.2011.05.005. In Press, Corrected Proof. [DOI] [PubMed] [Google Scholar]
- 50.Takeda A, Baffi JZ, Kleinman ME, et al. CCR3 is a target for age-related macular degeneration diagnosis and therapy. Nature. 2009;460:225–230. doi: 10.1038/nature08151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Y, Bedell M, Zhang K. Age-related macular degeneration: genetic and environmental factors of disease. Mol Interv. 2010;10:271–281. doi: 10.1124/mi.10.5.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seitsonen SP, Onkamo P, Peng G, et al. Multifactor effects and evidence of potential interaction between complement factor H Y402H and LOC387715 A69S in age-related macular degeneration. PLoS One. 2008;3:e3833. doi: 10.1371/journal.pone.0003833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmidt S, Hauser MA, Scott WK, et al. Cigarette Smoking Strongly Modifies the Association of LOC387715 and Age-Related Macular Degeneration. Am J Hum Genet. 2006;78:852–864. doi: 10.1086/503822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schaumberg DA, Hankinson SE, Guo Q, Rimm E, Hunter DJ. A prospective study of 2 major age-related macular degeneration susceptibility alleles and interactions with modifiable risk factors. Arch Ophthalmol. 2007;125:55–62. doi: 10.1001/archopht.125.1.55. [DOI] [PubMed] [Google Scholar]
- 55.Seddon JM, Reynolds R, Maller J, et al. Prediction model for prevalence and incidence of advanced age-related macular degeneration based on genetic, demographic, and environmental variables. Invest Ophthalmol Vis Sci. 2009;50:2044–2053. doi: 10.1167/iovs.08-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hughes AE, Orr N, Patterson C, et al. Neovascular age-related macular degeneration risk based on CFH, LOC387715/HTRA1, and smoking. PLoS Med. 2007;4:e355. doi: 10.1371/journal.pmed.0040355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakanishi H, Yamashiro K, Yamada R, et al. Joint effect of cigarette smoking and CFH and LOC387715/HTRA1 polymorphisms on polypoidal choroidal vasculopathy. Invest Ophthalmol Vis Sci. 2010;51:6183–6187. doi: 10.1167/iovs.09-4948. [DOI] [PubMed] [Google Scholar]
- 58.Tuo J, Ross RJ, Reed GF, et al. The HtrA1 promoter polymorphism, smoking, and age-related macular degeneration in multiple case-control samples. Ophthalmology. 2008;115:1891–1898. doi: 10.1016/j.ophtha.2008.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Francis PJ, Hamon SC, Ott J, Weleber RG, Klein ML. Polymorphisms in C2, CFB and C3 are associated with progression to advanced age related macular degeneration associated with visual loss. J Med Genet. 2009;46:300–307. doi: 10.1136/jmg.2008.062737. [DOI] [PubMed] [Google Scholar]
- 60.Conley YP, Jakobsdottir J, Mah T, et al. CFH, ELOVL4, PLEKHA1, and LOC387715 genes and susceptibility to Age-Related Maculopathy: AREDS and CHS cohorts and meta-analyses. Hum Mol Genet. 2006;15:3206–3218. doi: 10.1093/hmg/ddl396. [DOI] [PubMed] [Google Scholar]
- 61.Leveziel N, Souied EH, Richard F, et al. PLEKHA1-LOC387715-HTRA1 polymorphisms and exudative age-related macular degeneration in the French population. Mol Vis. 2007;13:2153–2159. [PubMed] [Google Scholar]
- 62.Kaur I, Katta S, Hussain A, et al. Variants in the 10q26 gene cluster (LOC387715 and HTRA1) exhibit enhanced risk of age-related macular degeneration along with CFH in Indian patients. Invest Ophthalmol Vis Sci. 2008;49:1771–1776. doi: 10.1167/iovs.07-0560. [DOI] [PubMed] [Google Scholar]
- 63.Gotoh N, Yamashiro K, Nakanishi H, et al. Haplotype analysis of the ARMS2/HTRA1 region in Japanese patients with typical neovascular age-related macular degeneration or polypoidal choroidal vasculopathy. Jpn J Ophthalmol. 2010;54:609–614. doi: 10.1007/s10384-010-0865-2. [DOI] [PubMed] [Google Scholar]
- 64.Chu J, Zhou CC, Lu N, Zhang X, Dong FT. Genetic variants in three genes and smoking show strong associations with susceptibility to exudative age-related macular degeneration in a Chinese population. Chin Med J (Engl) 2008;121:2525–2533. [PubMed] [Google Scholar]
- 65.Sng CC, Cackett PD, Yeo IY, et al. Toll-like receptor 3 polymorphism rs3775291 is not associated with choroidal neovascularization or polypoidal choroidal vasculopathy in Chinese subjects. Ophthalmic Res. 2011;45:191–196. doi: 10.1159/000321387. [DOI] [PubMed] [Google Scholar]
- 66.Conrad DF, Pinto D, Redon R, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Coin LJ, Asher JE, Walters RG, et al. cnvHap: an integrative population and haplotype-based multiplatform model of SNPs and CNVs. Nat Methods. 2010;7:541–546. doi: 10.1038/nmeth.1466. [DOI] [PubMed] [Google Scholar]
- 69.Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aitman TJ, Dong R, Vyse TJ, et al. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006;439:851–855. doi: 10.1038/nature04489. [DOI] [PubMed] [Google Scholar]
- 71.Hollox EJ, Huffmeier U, Zeeuwen PL, et al. Psoriasis is associated with increased beta-defensin genomic copy number. Nat Genet. 2008;40:23–25. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huffmeier U, Bergboer JG, Becker T, et al. Replication of LCE3C-LCE3B CNV as a risk factor for psoriasis and analysis of interaction with other genetic risk factors. J Invest Dermatol. 2010;130:979–984. doi: 10.1038/jid.2009.385. [DOI] [PubMed] [Google Scholar]
- 73.Ptacek T, Li X, Kelley JM, Edberg JC. Copy number variants in genetic susceptibility and severity of systemic lupus erythematosus. Cytogenet Genome Res. 2008;123:142–147. doi: 10.1159/000184701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang Y, Chung EK, Wu YL, et al. Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am J Hum Genet. 2007;80:1037–1054. doi: 10.1086/518257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Willcocks LC, Lyons PA, Clatworthy MR, et al. Copy number of FCGR3B, which is associated with systemic lupus erythematosus, correlates with protein expression and immune complex uptake. J Exp Med. 2008;205:1573–1582. doi: 10.1084/jem.20072413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu S, Yao L, Ding D, Zhu H. CCL3L1 copy number variation and susceptibility to HIV-1 infection: a meta-analysis. PLoS One. 2010;5:e15778. doi: 10.1371/journal.pone.0015778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Meyer KJ, Davis LK, Schindler EI, et al. Genome-wide analysis of copy number variants in age-related macular degeneration. Hum Genet. 2011;129:91–100. doi: 10.1007/s00439-010-0904-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jiang ST, Chiou YY, Wang E, et al. Essential role of nephrocystin in photoreceptor intraflagellar transport in mouse. Hum Mol Genet. 2009;18:1566–1577. doi: 10.1093/hmg/ddp068. [DOI] [PubMed] [Google Scholar]
- 79.Fu L, Garland D, Yang Z, et al. The R345W mutation in EFEMP1 is pathogenic and causes AMD-like deposits in mice. Hum Mol Genet. 2007;16:2411–2422. doi: 10.1093/hmg/ddm198. [DOI] [PubMed] [Google Scholar]
- 80.Narendran N, Guymer RH, Cain M, Baird PN. Analysis of the EFEMP1 gene in individuals and families with early onset drusen. Eye (Lond) 2005;19:11–15. doi: 10.1038/sj.eye.6701435. [DOI] [PubMed] [Google Scholar]
- 81.Hageman GS, Hancox LS, Taiber AJ, et al. Extended haplotypes in the complement factor H (CFH) and CFH-related (CFHR) family of genes protect against age-related macular degeneration: characterization, ethnic distribution and evolutionary implications. Ann Med. 2006;38:592–604. [PMC free article] [PubMed] [Google Scholar]
- 82.Schmid-Kubista KE, Tosakulwong N, Wu Y, et al. Contribution of copy number variation in the regulation of complement activation locus to development of age-related macular degeneration. Invest Ophthalmol Vis Sci. 2009;50:5070–5079. doi: 10.1167/iovs.09-3975. [DOI] [PubMed] [Google Scholar]
- 83.Fritsche LG, Lauer N, Hartmann A, et al. An imbalance of human complement regulatory proteins CFHR1, CFHR3 and factor H influences risk for age-related macular degeneration (AMD) Hum Mol Genet. 2010;19:4694–4704. doi: 10.1093/hmg/ddq399. [DOI] [PubMed] [Google Scholar]
- 84.Klein R, Klein BE, Knudtson MD, et al. Prevalence of age-related macular degeneration in 4 racial/ethnic groups in the multi-ethnic study of atherosclerosis. Ophthalmology. 2006;113:373–380. doi: 10.1016/j.ophtha.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 85.Polymeropoulos MH, Lavedan C, Leroy E, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 86.Singleton AB, Farrer M, Johnson J, et al. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 87.Hinds DA, Kloek AP, Jen M, Chen X, Frazer KA. Common deletions and SNPs are in linkage disequilibrium in the human genome. Nat Genet. 2006;38:82–85. doi: 10.1038/ng1695. [DOI] [PubMed] [Google Scholar]
- 88.Liu MM, Agron E, Chew E, et al. Copy Number Variations in Candidate Genes in Neovascular Age-Related Macular Degeneration. Invest Ophthalmol Vis Sci. 2011 doi: 10.1167/iovs.10-6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bressler SB, Munoz B, Solomon SD, West SK. Racial differences in the prevalence of age-related macular degeneration: the Salisbury Eye Evaluation (SEE) Project. Arch Ophthalmol. 2008;126:241–245. doi: 10.1001/archophthalmol.2007.53. [DOI] [PubMed] [Google Scholar]
- 90.Tuo J, Bojanowski CM, Zhou M, et al. Murine ccl2/cx3cr1 deficiency results in retinal lesions mimicking human age-related macular degeneration. Invest Ophthalmol Vis Sci. 2007;48:3827–3836. doi: 10.1167/iovs.07-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Age-Related Eye Disease Study Research G: A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E, beta carotene, and zinc for age-related macular degeneration and vision loss: AREDS report no. 8. Arch Ophthalmol. 2001;119:1417–1436. doi: 10.1001/archopht.119.10.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Smith TC, Lee L. Age related macular degeneration - new developments in treatment. Aust Fam Physician. 2007;36:359–361. [PubMed] [Google Scholar]
- 93.Parmeggiani F, Costagliola C, Gemmati D, et al. Coagulation gene predictors of photodynamic therapy for occult choroidal neovascularization in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2008;49:3100–3106. doi: 10.1167/iovs.07-1654. [DOI] [PubMed] [Google Scholar]
- 94.Parmeggiani F, Costagliola C, Gemmati D, et al. Predictive role of coagulation-balance gene polymorphisms in the efficacy of photodynamic therapy with verteporfin for classic choroidal neovascularization secondary to age-related macular degeneration. Pharmacogenet Genomics. 2007;17:1039–1046. doi: 10.1097/FPC.0b013e3282f12a4e. [DOI] [PubMed] [Google Scholar]
- 95.Parmeggiani F, Gemmati D, Costagliola C, Sebastiani A, Incorvaia C. Predictive role of C677T MTHFR polymorphism in variable efficacy of photodynamic therapy for neovascular age-related macular degeneration. Pharmacogenomics. 2009;10:81–95. doi: 10.2217/14622416.10.1.81. [DOI] [PubMed] [Google Scholar]
- 96.Lee AY, Raya AK, Kymes SM, Shiels A, Brantley MA., Jr Pharmacogenetics of complement factor H (Y402H) and treatment of exudative age-related macular degeneration with ranibizumab. Br J Ophthalmol. 2009;93:610–613. doi: 10.1136/bjo.2008.150995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee AY, Brantley MA., Jr CFH and LOC387715/ARMS2 genotypes and antioxidants and zinc therapy for age-related macular degeneration. Pharmacogenomics. 2008;9:1547–1550. doi: 10.2217/14622416.9.10.1547. [DOI] [PubMed] [Google Scholar]
- 98.Klein ML, Francis PJ, Rosner B, et al. CFH and LOC387715/ARMS2 genotypes and treatment with antioxidants and zinc for age-related macular degeneration. Ophthalmology. 2008;115:1019–1025. doi: 10.1016/j.ophtha.2008.01.036. [DOI] [PubMed] [Google Scholar]
- 99.Brantley MA, Jr, Fang AM, King JM, et al. Association of complement factor H and LOC387715 genotypes with response of exudative age-related macular degeneration to intravitreal bevacizumab. Ophthalmology. 2007;114:2168–2173. doi: 10.1016/j.ophtha.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 100.Nischler C, Oberkofler H, Ortner C, et al. Complement factor H Y402H gene polymorphism and response to intravitreal bevacizumab in exudative age-related macular degeneration. Acta Ophthalmol. 2011;89:344–349. doi: 10.1111/j.1755-3768.2010.02080.x. [DOI] [PubMed] [Google Scholar]
- 101.Blom AM, Kask L, Ramesh B, Hillarp A. Effects of zinc on factor I cofactor activity of C4b-binding protein and factor H. Arch Biochem Biophys. 2003;418:108–118. doi: 10.1016/j.abb.2003.08.018. [DOI] [PubMed] [Google Scholar]
- 102.Goverdhan SV, Hannan S, Newsom RB, et al. An analysis of the CFH Y402H genotype in AMD patients and controls from the UK, and response to PDT treatment. Eye (Lond) 2008;22:849–854. doi: 10.1038/sj.eye.6702830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Neale BM, Fagerness J, Reynolds R, et al. Genome-wide association study of advanced age-related macular degeneration identifies a role of the hepatic lipase gene (LIPC) Proc Natl Acad Sci U S A. 2010;107:7395–7400. doi: 10.1073/pnas.0912019107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Seitsonen SP, Jarvela IE, Meri S, et al. The effect of complement factor H Y402H polymorphism on the outcome of photodynamic therapy in age-related macular degeneration. Eur J Ophthalmol. 2007;17:943–949. doi: 10.1177/112067210701700612. [DOI] [PubMed] [Google Scholar]
- 105.Brantley MA, Jr, Edelstein SL, King JM, et al. Association of complement factor H and LOC387715 genotypes with response of exudative age-related macular degeneration to photodynamic therapy. Eye (Lond) 2009;23:626–631. doi: 10.1038/eye.2008.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.McKibbin M, Ali M, Bansal S, et al. CFH, VEGF and HTRA1 promoter genotype may influence the response to intravitreal ranibizumab therapy for neovascular age-related macular degeneration. Br J Ophthalmol. 2011 doi: 10.1136/bjo.2010.193680. [DOI] [PubMed] [Google Scholar]
- 107.Cho Y, Cao X, Shen D, et al. Evidence for enhanced tissue factor expression in age-related macular degeneration. Lab Invest. 2011;91:519–526. doi: 10.1038/labinvest.2010.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bell CG, Beck S. The epigenomic interface between genome and environment in common complex diseases. Brief Funct Genomics. 2010;9:477–485. doi: 10.1093/bfgp/elq026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fernandez-Morera JL, Calvanese V, Rodriguez-Rodero S, Menendez-Torre E, Fraga MF. Epigenetic regulation of the immune system in health and disease. Tissue Antigens. 2010;76:431–439. doi: 10.1111/j.1399-0039.2010.01587.x. [DOI] [PubMed] [Google Scholar]
- 110.Bentwich I, Avniel A, Karov Y, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 111.O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 112.Kutty RK, Nagineni CN, Samuel W, et al. Inflammatory cytokines regulate microRNA-155 expression in human retinal pigment epithelial cells by activating JAK/STAT pathway. Biochem Biophys Res Commun. 2010;402:390–395. doi: 10.1016/j.bbrc.2010.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kaneko H, Dridi S, Tarallo V, et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471:325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Damiani D, Alexander JJ, O’Rourke JR, et al. Dicer inactivation leads to progressive functional and structural degeneration of the mouse retina. J Neurosci. 2008;28:4878–4887. doi: 10.1523/JNEUROSCI.0828-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhou Q, Gallagher R, Ufret-Vincenty R, et al. Regulation of angiogenesis and choroidal neovascularization by members of microRNA-23~27~24 clusters. Proc Natl Acad Sci U S A. 2011;108:8287–8292. doi: 10.1073/pnas.1105254108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Baas DC, Ho L, Ennis S, et al. The complement component 5 gene and age-related macular degeneration. Ophthalmology. 2010;117:500–511. doi: 10.1016/j.ophtha.2009.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Goverdhan SV, Howell MW, Mullins RF, et al. Association of HLA class I and class II polymorphisms with age-related macular degeneration. Invest Ophthalmol Vis Sci. 2005;46:1726–1734. doi: 10.1167/iovs.04-0928. [DOI] [PubMed] [Google Scholar]
- 118.Despriet DD, Bergen AA, Merriam JE, et al. Comprehensive analysis of the candidate genes CCL2, CCR2, and TLR4 in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2008;49:364–371. doi: 10.1167/iovs.07-0656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kaur I, Hussain A, Hussain N, et al. Analysis of CFH, TLR4, and APOE polymorphism in India suggests the Tyr402His variant of CFH to be a global marker for age-related macular degeneration. Invest Ophthalmol Vis Sci. 2006;47:3729–3735. doi: 10.1167/iovs.05-1430. [DOI] [PubMed] [Google Scholar]