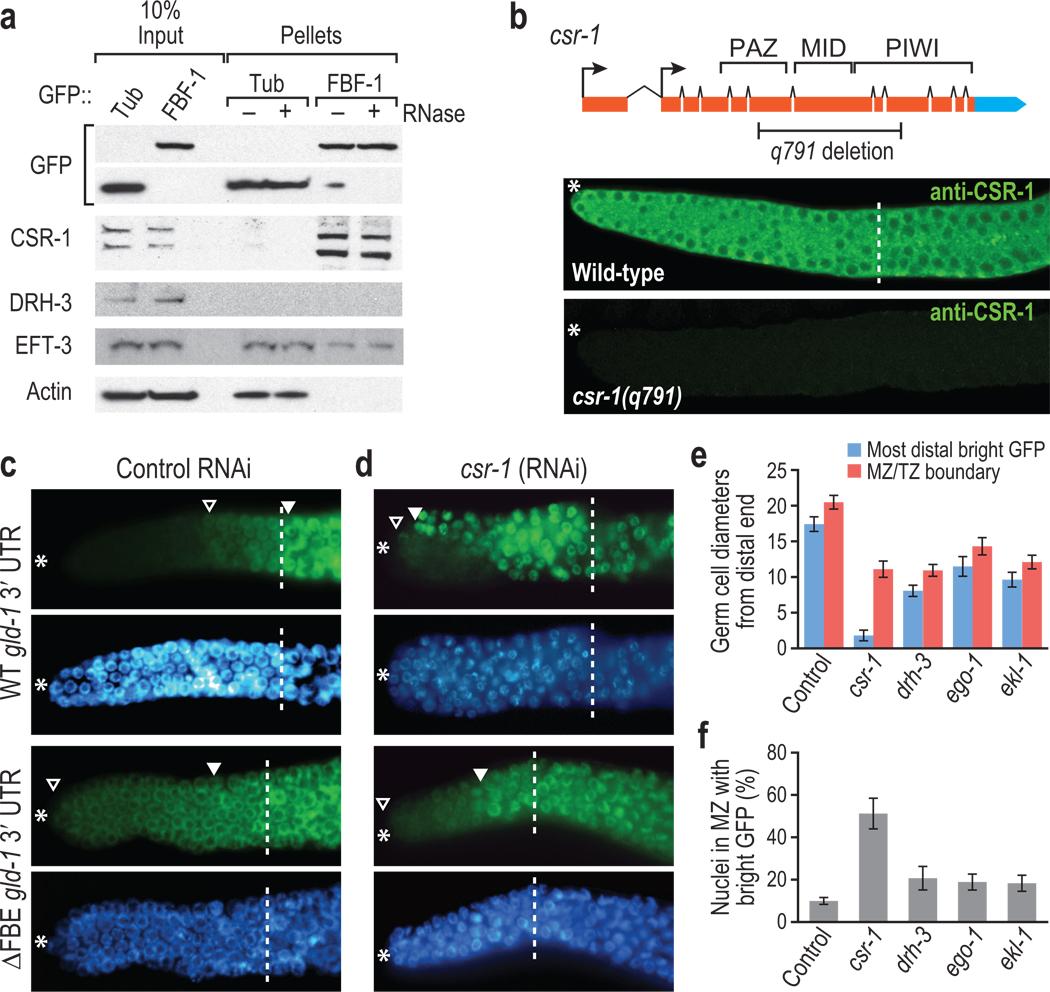
Figure 1.
FBF-1 binds CSR-1 to repress target mRNA. (a) FBF-1 co-IPs with CSR-1. Transgenic GFP::FBF-1 (FBF-1) was immunoprecipitated from adult hermaphrodites with or without RNase. Bound proteins (Pellets) were probed on Western blots for indicated proteins. CSR-1 and EFT-3 were both detected in FBF-1 IPs, whereas DRH-3 and actin were not. Two CSR-1 isoforms exist in vivo20,23, and both co-IP with FBF-1. CSR-1 did not co-IP with GFP::Tubulin (Tub). (b) CSR-1 is expressed in the mitotic zone. Top, diagram of csr-1 locus. The q791 deletion removes key domains resulting in a frameshift. Arrows, alternative promoter elements. Bottom: CSR-1 antibody23 stains the wild-type mitotic zone cytoplasm (*, distal end of the gonad; dashed line, mitotic zone/transition zone boundary), but not in a csr-1 mutant. (c, d) Adult germlines expressing GFP::H2B (green) under control of wild-type (WT) or FBE-lacking (ΔFBE) gld-1 3′ UTR; nuclei seen with DAPI (blue). Marked include: *, distal end; open triangle, distal-most GFP-positive cell; closed triangle, distal-most bright GFP; dashed line, mitotic zone/transition zone boundary. (c) Control RNAi, empty vector. GFP::H2B normally repressed in distal region (top panels), but expands into distal germ cells without FBEs (lower panels). (d) csr-1(RNAi). GFP::H2B expanded more distally than control in panel c when reporter harbors an FBE. (e) GFP::H2B extent after RNAi against genes indicated, scoring only sterile animals to ensure effective RNAi. Blue columns, most distal germ cell row with GFP::H2B-positive cell; red columns, mitotic zone/transition zone boundary. (f) Percent cells in mitotic zone with bright GFP::H2B.