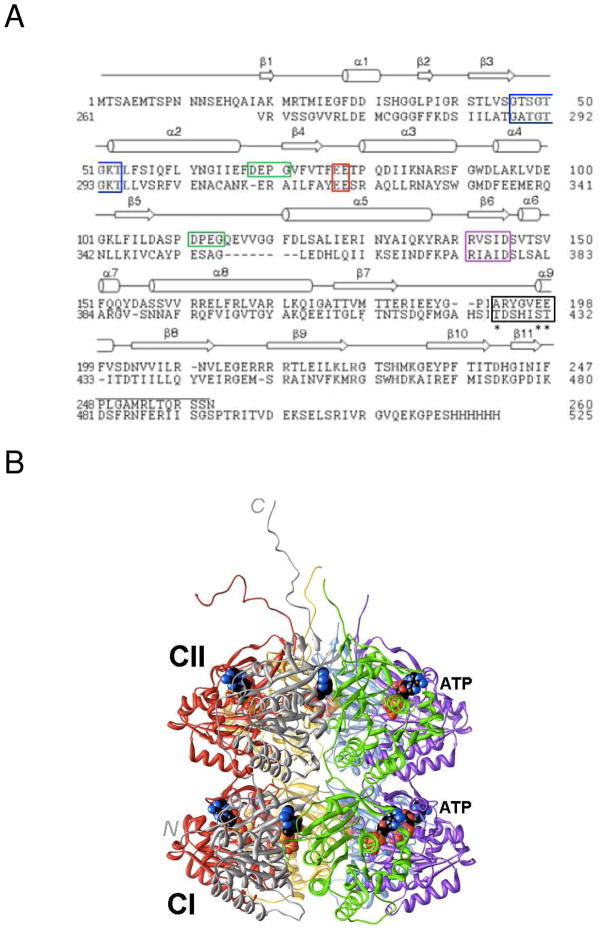
Figure 1.
Amino acid sequence and three-dimensional structure of S. elongatus KaiC. (A) Sequence alignment of the N- and C-terminal halves of KaiC (CI, top, and CII, bottom, respectively). Secondary structure elements as observed in the crystal structure28 are indicated above the primary structure and selected regions are boxed: Walker A motif (P-loop GXXXXGKT, blue), truncated Walker B motif (RXXXD, purple), sequence conserved in some GTP-binding proteins (DXXG, green), putative catalytic carboxylate(s) (red), and phosphorylation loop (black, phosphorylation sites in CII, T432, S431, and T426 are marked with an asterisk). (B) Crystal structure of full-length KaiC (PDB ID 3DVL; http://www.rcsb.org).12,28 Hexamer subunits A, B, C, D, E and F are colored gray, green, magenta, blue, yellow and red, respectively. ATP molecules bound between subunits are shown in space filling mode with carbon atoms colored in black. At the N-terminal end of CI, residues 1–13 are missing. C-terminal peptide tails of CII (489–519) that serve as the binding site for KaiA12,49 were only traced completely for subunits A and F.