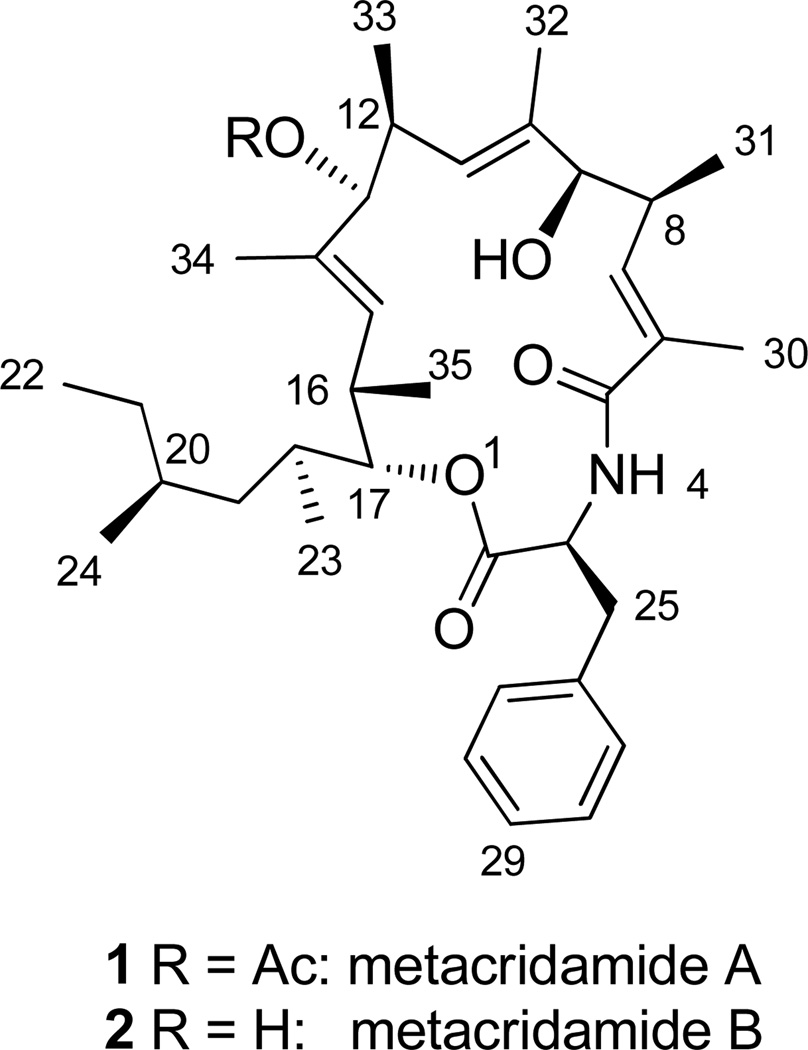
Abstract
Metarhizium acridum, an entomopathogenic fungus, has been commercialized and used successfully for biocontrol of grasshopper pests in Africa and Australia. Its conidia produce two novel 17-membered macrocycles, metacridamides A (1) and B (2), which consist of a Phe unit condensed with a nonaketide. Planar structures were elucidated by a combination of mass spectrometric and NMR techniques. Following hydrolysis of 1, chiral amino acid analysis assigned the L-configuration to the Phe unit. A crystal structure established the absolute configuration of the eight remaining stereogenic centers in 1. Metacridamide A (1) showed cytotoxicity to three cancer lines with IC50s of 6.2, 11.0, and 10.8 µM against Caco-2 (epithelial colorectal adenocarcinoma), MCF-7 (breast cancer), and HepG2/C3A (hepatoma) cell lines, respectively. In addition, metacridamide B (2) had an IC50 of 18.2 µM against HepG2/C3A, although it was inactive at 100 µM against Caco-2 and MCF-7. Neither analogue showed antimicrobial, phytotoxic, or insecticidal activity.
Metarhizium acridum (Bischoff, Humber, and Rehner) (= M. anisopliae var. acridum) (Cordicepitaceae) is an entomopathogenic fungus with a host-range limited to grasshoppers in the orthopteran family Acrididae.1 The conidia of M. acridum constitute the active ingredient of commercially produced biopesticides, marketed as Green Muscle® and Green Guard®, that are used for control of migratory locusts in Africa and Australia, respectively.2 Application of Green Muscle® against Red Locust over 10,000 hectares of Iku-Kitavu National Park Tanzania in 2009 was credited with curtailing a potentially devastating locust outbreak and averting extensive crop losses that could have affected the food security of ca. 15 million people in the region.3
We have an ongoing interest in the secondary metabolites of biocontrol fungi in the genus Metarhizium4,5 with a focus on the possibility that they contribute to pathogenicity and/or virulence.6,7 The widespread dissemination of M. acridum for insect biocontrol compels continued study of its basic biology to gain insights that can lead to improved efficacy and shelf-life of the formulated product8 and to expose potential chemical risks associated with use of these fungi as biopesticides.5
We recently identified the serinocyclins, cyclic heptapeptides produced by conidia of M. acridum and M. robertsii, and identified the gene responsible for their biosynthesis in the latter.4,6 The discovery of the serinocyclins from conidia as opposed to mycelium or broth from a liquid culture revealed an underexploited source of natural product diversity in Metarhizium fungi. Herein we describe the isolation, characterization, and biological activity of metacridamides A (1) and B (2), two novel cytotoxic macrocycles from the conidia of M. acridum.
RESULTS AND DISCUSSION
Low resolution positive ion ESIMS analyses of crude hexane extracts of conidia of M. acridum (ARSEF #3341) showed a predominant signal at m/z 632 consistent with the sodium adduct [M+Na]+ of a compound with a molecular weight of 609 da. Negative ion spectra showing peaks at m/z 608 [M-H]−, 644/646 [M+Cl]−, and 654 [M+HCOO]− supported this conclusion. As there were no other candidates with a unit mass of 609 Da previously reported from M. acridum, the compound was earmarked for isolation and characterization.
RP HPLC of the crude hexane extracts produced chromatograms with two major peaks at tR = 6.50 and 5.53 min with the latter as the minor peak. Preparative RP HPLC led to the isolation of the major compound (1) at tR = 6.50 min which was shown by ESIMS to have a molecular mass of 609 Da. The compound (2) giving rise to the minor peak gave a prominent signal in positive ion ESIMS at m/z 590 consistent with the [M+Na]+ of a compound with a molecular mass of 567 Da. The 42 Da difference between the pseudomolecular ions of (1) and (2) suggested an acetylated compound and its deacetyl analogue. MS analysis of a sample of (1) dissolved in CH3OD for deuterium exchange showed [M+Na]+ and [M+K]+ at m/z 634 and 650 respectively, indicating the presence of 2 exchangeable protons.
1H and COSY NMR spectra of 1 (Table 1) indicated the presence of 53 non-exchangeable protons, including one methyl triplet, eight methyl doublets (three of which showed coupling constants of < 1 Hz suggesting a long-range (4J) coupling), and a methyl singlet. Three well-resolved doublets resonated at δH 6.03, 5.11, and 5.02, suggesting alkenyl protons. Five protons in the aromatic region suggested a monosubstituted phenyl group.
Table 1.
NMR Spectroscopic Data for Metacridamides A (1) and B (2)
| metacridamide A (1)a |
metacridamide B (2)b |
|||||
|---|---|---|---|---|---|---|
| position | δC, mult.c | δH (J in Hz)d | HMBCe | δCf | δH (J in Hz)d | HMBCe |
| 2 | 170.9, qC | 173.1 | ||||
| 3 | 53.0, CH | 4.94, m | 2, 5, 25 | 54.4 | 4.85 br, s | |
| 4 | 5.91, d (8.6) | 2g, 3, 5, 25 | 8.21, d (9.1) | 3, 5 | ||
| 5 | 168.7, qC | 172.6 | ||||
| 6 | 132.7, qC | 132.5 | ||||
| 7 | 133.7, CH | 6.03, dd (9.5, 1.4) | 5, 8g, 9, 30, 31 | 136.9 | 6.15, d (9.5) | 5, 30 |
| 8 | 35.5, CH | 2.84, m | 6, 7, 9g, 31 | 36.8 | 2.81h, m | 6, 7, 31 |
| 9 | 78.3, CH | 3.95, s | 7, 8, 10, 11, 31, 32, 33g,i | 79.3 | 3.94, s | 7, 8, 10, 11, 31, 32, 33g,i |
| 10 | 137.1, qC | 136.3 | ||||
| 11 | 124.3, CH | 5.02, m | 9, 10g, 12g, 13g, 32, 33 | 127.4 | 5.26, d (10.5) | 9, 32, 33 |
| 12 | 36.6, CH | 2.83, m | 9g,i, 10, 11, 13, 33 | 40.7 | 2.73, ddd (10.5, 7.0, 3.6) | 10, 11, 13, 33 |
| 13 | 81.4, CH | 4.92, m | 11, 12, 14, 15, 33, 34, Ac C=O | 81.0 | 3.84, d (10.2) | 11, 12, 14, 15, 33, 34 |
| 14 | 130.9, qC | 136.0 | ||||
| 15 | 131.0, CH | 5.11, dd (10.0) | 13, 16, 17, 34, 35g | 130.3 | 5.12, d (10.2) | 13, 16, 17, 34, 35 |
| 16 | 33.5, CH | 2.71, m | 14, 15, 35 | 35.5 | 2.81h, m | 14, 15, 35 |
| 17 | 82.7, CH | 4.61, dd (8.8, 2.3) | 2, 15, 16, 18, 19, 23, 35 | 83.7 | 4.68, dd (8.0, 1, 8) | 2, 15, 16g, 18g, 19, 23, 35 |
| 18 | 33.7, CH | 1.61, m | 19, 20, 23 | 35.2 | 1.70, m | 19g |
| 19 | 40.2, CH2 | 1.10, ddd (13.3, 9.5, 3.8) | 17, 18, 20, 21, 23, 24 | 42.3 | 1.29, m | 18, 20, 21, 23, 24 |
| 0.84, m | 17, 18, 20, 21, 23, 24 | 0.91, m | 18, 20, 21, 23, 24 | |||
| 20 | 31.2, CH | 1.38, m | 33.0 | 1.43, m | ||
| 21 | 27.6, CH2 | 1.28, m | 19, 20, 22g, 24 | 29.4 | 1.39, m | 19g, 20, 22g, 24 |
| 0.84, m | 19, 20, 22, 24 | 0.96, m | 19g, 20, 22, 24 | |||
| 22 | 10.8, CH3 | 0.74, t (7.4) | 20, 21 | 11.7 | 0.85, t (7.4) | 20, 21 |
| 23 | 16.0, CH3 | 0.75, d (6.7) | 17, 18, 19 | 16.3 | 0.77, d (6.7) | 17, 18, 19 |
| 24 | 20.5, CH3 | 0.81, d (6.6) | 19, 20, 21 | 20.8 | 0.88 | 19, 20, 21 |
| 25 | 36.5, CH2 | 3.27, dd (14.0, 6.1) | 2, 3, 26, 27 | 37.2 | 3.33, dd (14.1, 5.4) | 2, 3, 26, 27 |
| 3.20, dd (14.0, 5.8) | 2, 3, 26, 27 | 2.95, dd (14.1, 10.2) | 2, 3, 26, 27 | |||
| 26 | 137.0, qC | 139.5 | ||||
| 27, 27’ | 129.8, CH | 7.20, m | 25, 29 | 130.4 | 7.233, m | 25, 29 |
| 28, 28’ | 128.6, CH | 7.27, m | 26, 27g | 129.4 | 7.235, m | 26 |
| 29 | 127.0, CH | 7.21, m | 27, 28g | 127.6 | 7.17, m | 27, 28g |
| 30 | 13.1, CH3 | 1.83, d (1.4) | 5, 6, 7, 8g,j, 9g,i, 31g,i | 13.6 | 1.65, d (1.4) | 5, 6, 7, 31g,i |
| 31 | 19.4, CH3 | 1.19, d (7.0) | 7, 8, 9 | 19.3 | 1.18, d (7.0) | 6g,j, 7, 8, 9 |
| 32 | 15.0, CH3 | 1.61, m | 8g,j, 9, 10, 11, 12g,j | 15.2 | 1.60, d (0.6) | 9, 10, 11 |
| 33 | 16.1, CH3 | 0.92, d (7.0) | 11, 12, 13, 34g,i | 16.1 | 0.93, d (7.0) | 11, 12, 13, 34g,i |
| 34 | 13.6, CH3 | 1.63, d (1.3) | 12g,j, 13, 14, 15, 16g,j, 35i | 13.7 | 1.67, d (1.3) | 12g,j, 13, 14, 15, 35g,i |
| 35 | 18.2, CH3 | 0.67, d (6.9) | 15, 16, 17 | 19.8 | 1.02, d (6.9) | 15, 16, 17 |
| Ac C=O | 169.8, qC | |||||
| Ac CH3 | 21.4, CH3 | 2.04, s | Ac C=O, 13g,i | |||
CDCl3.
CD3OD (assignments and correlations for H4 obtained in CD3OH).
150MHz.
600 MHz.
HMBC correlations, optimized for 8 MHz, are from proton(s) stated to the indicated carbon.
125 MHz.
Weak crosspeak.
Overlapping signals.
5-bond correlation (Figures S16, S32).
4-bond correlation.
The 13C spectrum showed signals for 37 carbons. HSQC data corroborated the presence of 10 methyl carbons (δC 10.8–21.4) and revealed three sp3 methylenes, six sp3 methine carbons, three oxymethines, three alkenyl methines, and five aromatic methine carbons for a total of thirty carbons with attached protons. By process of elimination seven quaternary carbon signals were identified from the 13C spectrum, three in the carbonyl region and four in the 131–137 ppm range consistent with alkenyl or aromatic carbons.
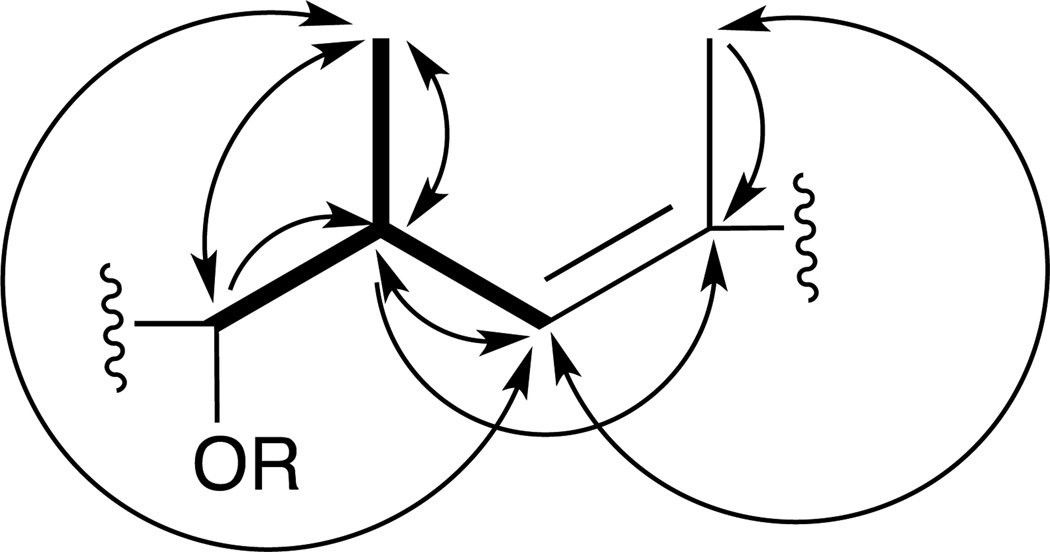
COSY and HSQC spectra showed three conspicuously similar spin systems in each of which a complex multiplet in the 2.6–2.8 ppm range coupled with (1) a methyl doublet in the 0.6–1.2 ppm range, (2) a proton in the 3.9–4.9 ppm range, suggesting an oxygenated methine carbon, and (3) a doublet in the 5.1–6.1 ppm range, suggesting an olefinic methine proton vicinal to a quaternary alkenyl carbon. In HMBC spectra the alkenyl proton either did not correlate or showed only a weak 2J correlation to a quaternary olefinic carbon (δC 130.9–137.1). However, this carbon was correlated with another set of methyl protons (δH 1.60–1.83) as well as with a methine proton (δH 2.71–2.84) in all three units. The correlations associated with these three similar units indicated the 6-carbon substructure depicted in Figure 1.
Figure 1.
6-carbon unit consistent with COSY, HSQC, and HMBC spectra of 1. Bolded bonds indicate COSY network. Arrows indicate HMBC correlations (carbon detected by indicated proton). Double-headed arrows indicate reciprocal HMBC correlations.
The spin system of a Phe unit was revealed by a proton doublet signal at δH 5.91 that showed no attached carbon in the HSQC spectrum. The assignment of this signal to an amide proton was supported by its COSY correlation with an α-methine proton at δH 4.94, which in turn coupled with two geminal β-protons at δH 3.27 and 3.20. HMBC spectra showed correlations between the β(H25) protons and the γ (C26) and δ (C27) carbons in the phenyl ring. In addition, in HMBC spectra, both the α–(H3) and amide protons correlated with both the carbonyl carbon (C2) of the Phe unit and its acylating carbonyl carbon (C5).
A COSY correlation between the methyl triplet at δH 0.74 (H22) and a pair of methylene protons at δH 1.28 and 0.84 (H21a,b) provided an entry point for deciphering a 7-carbon aliphatic group that took shape from NMR data as a 4-methylhexane 2-yl group.
HMQC and HMBC correlations readily confirmed the acetate group predicted from MS data as the R-group in one of the units depicted in Figure 1. In HMBC spectra the acetate methyl protons (δH 2.04) correlated with the acetate carbonyl carbon (δC 169.8) as well as the methine proton at δH 4.92, which HSQC data indicated is attached to the oxygenated carbon at δC 81.4 (C13). The oxygenated methine proton at δH 3.95 (H9) correlated with the quaternary alkenyl carbon at δC 137.1 (C10) and its adjacent methyl carbon at δC 15.0 (C32). These correlations thus linked the oxymethine carbon of a second unit with the quaternary alkenyl carbon of the unit carrying the acetyl group. An analogous set of correlations linked the acetylated carbon (C13) with the alkenyl quaternary carbon (C14) of the third unit. HMBC correlations were observed between the oxymethine proton at δ 4.62 of this third unit and the carbonyl carbon of the Phe group (C2) at δC 170.9 as well as with the methine carbon of the 4 methyl-hexan-2-yl unit (C18) at δC 33.7. The COSY correlation of the C18 methine proton at δH 1.61 with the methine at δ 4.61 and the 3J HMBC correlation H23→C17 and its complement H17→C23 confirmed the linkage of C17 to C18 of the 2-yl of the 4 methyl-hexan-2-yl group.
The HMBC correlation between the H30 methyl protons (δH 1.83) and the C5 carbonyl carbon, which acylates the amide nitrogen of the Phe unit, as well as the correlation between the H7 olefinic proton (δH 6.03) and this carbonyl, established the bond between the quaternary alkenyl carbon (C6) and C5 and, thus, completed the 17-member macrocycle of 1 (Chart 1; Table 1).
Chart 1.
HRESIMS data for 1 indicating the molecular formula C37H55NO6 and an index of unsaturation of 11 were consistent with the planar structure deduced from NMR data and also allowed for two deuterium-exchanged protons, the amide proton and the hydroxyl proton on the C9 oxygen.
Direct chiral amino acid analysis by Cu+2 ligand-exchange chromatography indicated the presence of L-Phe in 1. The absolute configuration of the Phe unit of 1 was identified by co-chromatography of the hydrolysis product of 1 with a standard mixture consisting of D- and L- Phe (Figure S35).
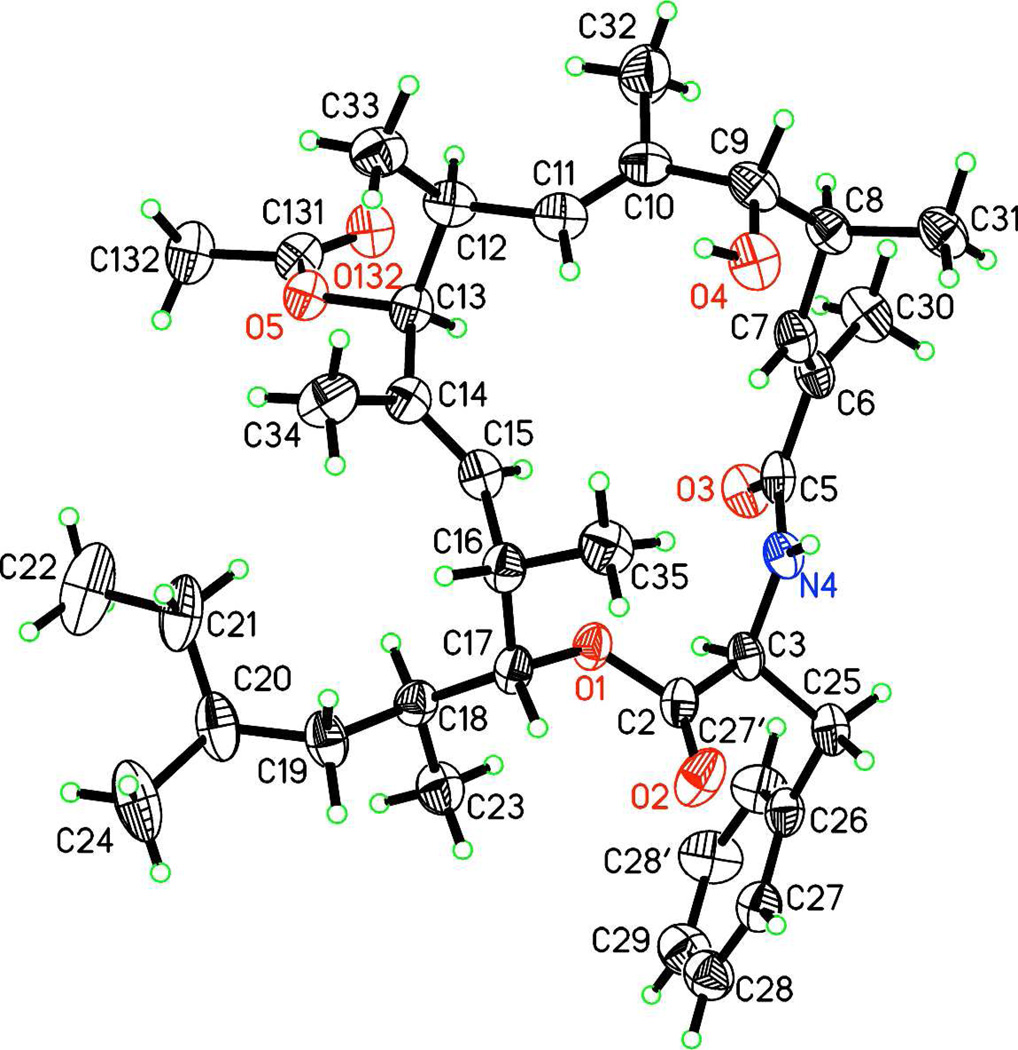
The crystal structure of 1 was determined using X-ray diffraction and standard techniques (Figure 2).10 This structure, combined with the results of chiral amino acid analysis, established the absolute configuration of all nine stereocenters as well as the geometry of the three double-bonds in 1 as 3S,6E,8S,9R,10E,12S,13S,14E,16S,17R.
Figure 2.
Crystal structure of 1, with 50% probability thermal ellipsoids for non-H atoms.
HRESIMS data for 2 were consistent with the molecular formula C37H55NO6 confirming that 2 differed from 1 by the replacement of the acetyl group in 1 with a hydroxyl group. NMR data for 2 lacked all signals and correlations associated with the acetyl group of 1. Because 2 was isolated in much smaller quantities than 1, attempts were made to conserve material for bioassays by finding an aprotic NMR solvent other than d6-DMSO. However, 2 was insoluble in all common low-boiling aprotic solvents tried (CDCl3, CD2Cl2, CD3CN, d6 acetone) and was, therefore, dissolved in CD3OD for NMR studies. As expected, the amide proton of the Phe unit of 2 was not detected in NMR experiments in CD3OD because of deuterium exchange. An abbreviated series of experiments using CD3OH revealed the amide proton (δH 8.21) and its correlations (Table 1, Figures S29, S33). Most of the key correlations observed in 2-D spectra of 1 were also observed for 2 (Table 1). The absolute configuration of 2 was deduced by analogy to 1.
The transannular ROESY correlations between the C7 olefinic methine proton and the C35 methyls were observed in the spectra of both 1 (Figure S17, S36) and 2 (Figure S34). In this respect the solution structures were consistent with the crystal structure of 1 in which the H35 methyl group is the only one of the ten in the molecule that projects into the interior of the macrocycle and its protons can approach within 3 A of the H7 proton.
Weak but clearly resolved ROESY correlations to the H27 (δ-Phe) and H28 (ε-Phe) protons from H35, H31 and H7 were observed for 1 (but not for 2 possibly because the sample was much more dilute). In the crystal structure all interatomic distances involved in these correlations are greater than 6 A and thus, would be unlikely to produce a ROESY crosspeak (Figure S37). Using a computer generated 3-D model (Chem-3-D® Pro) of the crystal structure of 1 all the relevant interatomic distances can be brought into the 2–4 A range by rotations around the C3-C25 (Phe α-β) and C-25-C26 (Phe β-γ) bonds (Figure S38). Thus, the ROESY data suggest that these bonds rotate freely in solution.
Metacridamide A (1) showed no antimicrobial activity at 100 µg/disk, no insecticidal activity against Drosophila melanogaster at a 100 µg dosage, no larvicidal activity against Aedes aegypti at 100 µg/ml, and no phytotoxicity activity at 50 µg/mL against lettuce seedlings. However, it did show moderate cytotoxicity against three cancer lines with IC50s of 6.2, 11.0, and 10.8 µM against Caco-2 (epithelial colorectal adenocarcinoma), MCF-7 (breast cancer), and HepG2/C3A (hepatoma) cell lines, respectively. In addition, metacridamide B (2) had an IC50 of 18.2 µM against HepG2/C3A, although it was inactive at 100 µM against Caco-2 and MCF-7.
The uninterrupted 18-carbon chain starting at C5 and ending at C22 in both 1 and 2 bears the hallmarks of a polyketide.11 The carbon skeleton, as well as all of the modifications of this putative nonaketide moiety, can be accounted for as products of the well-characterized functionalizing enzymes associated with Type I fungal polyketide synthases (ketoreductase, dehydratase, enol-reductase, and a C-methyl transferase employing S-adenosyl methionine (SAM) as a source of methyl groups). The acetyl group at C13 of 1 could also be the product of a closely linked or heterologous acetyl transferase.
It seems likely that the Phe group, which forms the 17-membered macrocycle by an ester linkage to O1 at its C-terminal and an amide linkage to C5 at its N-terminal, is incorporated by a nonribosomal peptide synthetase (NRPS).12 Hybrid PKS-NRPS genes are widespread in fungi.7,13–15 We expect that recently published genome sequence data16 will facilitate identification of hybrid synthetases in M. acridum and that a functional analysis using a gene knockout approach will identify the gene responsible for the metacridamide biosynthetic pathway.
A search of the literature revealed two examples of recently discovered fungal natural products whose structures and patterns of oxygenated carbons and alkenyl groups suggest biosynthetic pathways that are similar, if not homologous, to those of the metacridamides. A compound from the patent literature derived from an unidentified soil fungus combines a Phe unit with a heptaketide.17 The torrubiellutins combine an N-methylated Phe unit with a presumptive hexaketide.18 Intriguingly, the torrubiellutins, like the metacridamides, derive from an entomopathogenic fungus. Other PKS-NRPS hybrid macrocycles incorporating a Phe unit include hapalosin 19 and the cytochalasins.20 The chondropsins,21 although lacking a Phe unit, contain three iterations of the substructure depicted in Figure 1, two of which are contiguous.
As with many microbial natural products the biological activity of the metacridamides was revealed in this study in assays that can have little if any relevance to the context in which these compounds evolved. There is no obvious adaptive rationale for the cytotoxicity of a compound produced by the spores of fungal pathogens of insects. There are now many compounds known from entomopathogenic Metarhizium fungi with a wide range of biological activities ascribed to them,22 but virtually nothing is known about how they function for the organisms that produce them. The looming challenge is to advance our understanding of the biology of these compounds to a point in line with our ability to define them chemically.
EXPERIMENTAL SECTION
General Experimental Procedures
Optical rotations were measured at 25 °C in MeOH on a Rudolph Autopol IV polarimeter at 589 nm with a 100 mm cell. 1H, 13C, gCOSY, DQCOSY, ROESY, HSQCAD, and gHMBCAD NMR experiments were carried out on Varian Inova 400, 500 or 600 spectrometers using either HSW, DBG broad-band decoupler, 5 mm switchable, or HCN inverse detection probes. Pulse sequences were provided in Varian VnmrJ 2.2D/Chempack 4.1. HSQC and HMBC spectra were acquired at 600 MHz (1H-dimension), and were optimized for 1JCH = 150, 190 Hz and nJCH = 8.0, 5.0, respectively. ROESY spectra were acquired with a mixing time of 0.2 s.
NMR data were recorded for samples dissolved in CDCl3, CD3OD, or CD3OH. 1H chemical shifts were referenced to the singlet at δH 7.26 or center of the residual CHD2OD quintuplet at δH 3.31. 13C chemical shifts were referenced to the center of the 13CD3OD septuplet at δC 49.15 or the CDCl3 triplet at δC 77.16. Low resolution ESI mass spectra were acquired by infusion of methanolic solutions at 5 µL/min by a syringe pump (Harvard Apparatus) into a Micromass ZMD 4000 spectrometer. High resolution ESI mass spectra were similarly acquired in positive ion mode on a Waters XEVO-G2 spectrometer using lock mass correction with leu-enkephalin as the external lock-mass standard.
Fungal Material and Culture Conditions
Conidia of M. acridum were produced using a published protocol.8 Briefly, cryogenically preserved mycelial material (ARSEF #3341 and #7486, both derived from IMI-330189 which was isolated from an unidentified grasshopper in Niger) was used to inoculate 100 mL liquid cultures (20 g glucose, 20 g yeast extract/L H2O) in 250 mL erlenmeyer flasks which were shaken at 150 rpm for 3 days at 20±3 °C. The liquid starter culture was then used to inoculate solid cultures on barley (100 g rolled barley flakes moistened with 60 mL H2O) in autoclavable perforated polyethylene "spawn bags" (Myco Supply).
Extraction and Isolation
A simple extraction protocol was developed to optimize the yield of 1 and 2. Conidia were suspended in hexane (200 mL/13 g conidia) in 250 mL polypropylene centrifuge bottles. The suspensions were vortexed for 30 seconds, sonicated for 30 minutes, and then centrifuged to pellet conidia. The supernatants were decanted and concentrated in vacuo. Ca. 160 g of conidia afforded ca. 156 mg of a whitish solid residue. This was then vortexed vigorously in MeOH and the supernatant was filtered and chromatographed by semi-preparative reversed-phase HPLC (Phenomenex Prodigy ODS3; 10×250 mm; 5 μm; 100 Å) eluting at 4 mL/min using an isocratic mixture of MeOH (95) : H2O (5) and UV detection (λ = 215 nm). Fractions containing 1 and 2 were polished using the same HPLC column with a mobile phase consisting of MeCN(9) : H2O (1). The mixture furnished 1 (18 mg) and 2 (6 mg) of >99% purity (tR = 6.50 and 5.53 min, respectively).
Metacridamide A (1)
colorless plates (MeOH); [α]26D: −10.0 (c 1.20, MeOH); UV (MeOH) λmax (log ε) 233 (3.34). 1H, 13C NMR, HSQC, HMBC, ROESY data, see Table 1 and the Supporting Information; HRESIMS; obsd m/z 632.3925 [M+Na]+; calcd for C37H55NO6 + Na, 632.3927, HPLC tR = 6.50 min.
Metacridamide B (2)
colorless oil; [α]24D: +1.3 (c 0.34, MeOH); UV (MeOH) λmax (log ε) 236 (3.36). 1H, 13C NMR, HSQC, HMBC, ROESY data, see Table 1 and the Supporting Information; HRESIMS; obsd m/z 590.3820 [M+Na]+; calcd for C35H53NO5 + Na; 590.3821, HPLC tR = 5.53 min.
Chiral amino acid analysis
A 0.7 mg sample of 1 was hydrolyzed in a 1:1 mixture of 12M HCl and propionic acid at 110 °C for 15 h and then dried in vacuo to yield a white solid and a brown gum. The latter was removed by trituration with 0.4 mL dichloromethane. The solid white residue was taken up in 0.25 mL H2O. 10 µL aliquots were co-chromatographed with standard D- and L-Phe (Sigma-Aldrich) using a Cu+2 ligand-exchange HPLC column (Phenomenex Chirex (D)-penicillamine; 4.6×250 mm; 5μm) eluted at 1 mL/min with 2mM CuSO4 in MeOH:H2O (3:7); detection by UV absorption at 254 nm.
X-ray Crystallography of 1
X-ray crystallographic data for compound 1 were collected at the F1 beamline at the Cornell High Energy Synchrotron Source using an ADSC Quantum 270 CCD detector that was offset to allow collection of data up to a resolution of 0.87 Å.23
A single crystal with dimensions of 100×10×5 μm was obtained by dissolving the sample in MeOH, adding 2% HCOOH dropwise to induce precipitation, clearing the solution with another drop of HCOOH and then slowly evaporating the sample at 20 °C. The crystal was then mounted on a Mitegen micro mesh mount24 using a small amount of highly viscose NVH oil (Hampton Research). Data were collected at 100 K at a wavelength λ = 0.9177 Å. Using a microcrystal setup with focusing glass capillary (focal spot diameter 18 μm) 75 φ-wedges 5 deg. each with 4 sec. exposure time were recorded.
Data were integrated with XDS and scaled with XSCALE.25,26 15274 observations were merged into 4991 unique reflections (Space Group P212121, Rint = 4.32%, 96.4 % completeness).
The crystal structure was solved by direct methods (SHELXS-97) and refined using full-matrix least-squares methods on F2 (SHELXL-97).27 Hydrogen atoms were generated geometrically and included in the refinement in the riding model approximation. All non-hydrogen atoms were refined anisotropically. Final results for 398 parameters: R1 = 5.31%, wR2 = 13.15%, GooF = 1.056.
Bioassays
Metacridamide A (1) was tested against a panel of bacterial and fungal targets in disk-diffusion assays. Escherichia coli and Bacillus cereus were grown on nutrient agar, Colletotrichum acutatum, Botrytis cinerea, and Beauveria bassiana (ARSEF #8354) on potato dextrose agar, and Saccharomyces cerevisiae on 1% yeast extract, 2% peptone, 1.5% agar, supplemented with either 2% dextrose or 2% glycerol. Plates were spread with 0.6 mL aliquots of a liquid bacterial culture grown for 24 h in nutrient broth, 0.6 mL of a fungal conidial suspension (ca. 1×106 conidia/mL H2O), or with 0.5 mL of a yeast suspension in H2O adjusted to OD600 = 0.5. Inoculated plates were allowed to dry for 1 h. Test samples were applied to 7-mm filter paper disks in 10 μL MeOH. Disks were air-dried for 1 h and then placed on assay plates. Cultures were monitored for zones of inhibition every 24 h for 4 days. Tetracycline and filipin were used at 10 μg/disk as positive controls for the bacterial and fungal targets, respectively. MeOH was used at 10 μL/disk as a negative control.
Using previously published methods 1 was also tested for phytotoxicity in a lettuce seedling elongation assay,28 and for insecticidal activity against Drosophila melanogaster adults29 and Aedes aegypti larvae.4
Cytotoxicity Assays
Metacridamides A and B (1 and 2) were assayed for toxicity against Caco-2 (epithelial colorectal adenocarcinoma), MCF-7 (breast cancer), and HepG2/C3A (hepatoma) cell lines using the XTT assay30 with the XTT Cell Proliferation KIT II (Roche Applied Science).
Supplementary Material
ACKNOWLEDGEMENT
We gratefully acknowledge R. Vaughan and Mary Bodis (USDA-ARS), J.Y. Maeng, J. Zuk (Cornell), and I. Keresztes (Cornell Chemistry and Biology) for technical assistance and I. Molnar (University of Arizona) for helpful discussion. The L. Harrington and B. Lazzaro laboratories (Cornell Entomology) kindly provided insects for bioassays. X-ray diffraction data were collected at the Cornell High Energy Synchrotron Source, which is supported by the National Science Foundation under award DMR-0225180, using the MacCHESS facility, supported by award RR-01646 from the National Institutes of Health. Mention of a trademark, proprietary product, or vendor does not constitute a guarantee or warranty of the product by the U.S. Department of Agriculture and does not imply its approval to the exclusion of other products or vendors that may also be suitable.
Footnotes
ASSOCIATED CONTENT
Supporting Information. 1H, 13C, COSY, HSQC, HMBC, and ROESY spectra of 1 and 2, chiral HPLC amino acid analysis data for 1, 3-D models of 1 with interatomic distances for selected protons, tabulated X-ray data for 1, and tabulated interatomic distance data for selected proton pairs of 1. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES AND NOTES
- 1.Bischoff JF, Rehner SA, Humber RA. Mycologia. 2009;101:512–530. doi: 10.3852/07-202. [DOI] [PubMed] [Google Scholar]
- 2.Lomer CJ, Bateman RP, Johnson DL, Langewald J, Thomas MB. Annu. Rev. Entomol. 2001;46:667–702. doi: 10.1146/annurev.ento.46.1.667. [DOI] [PubMed] [Google Scholar]
- 3.Anonymous New Agriculturalist. 2009 September; http://www.new-ag.info/developments/devItem.php?a=943.
- 4.Krasnoff SB, Keresztes I, Gillilan RE, Szebenyi DME, Donzelli BGG, Churchill ACL, Gibson DM. J. Nat. Prod. 2007;70:1919–1924. doi: 10.1021/np070407i. [DOI] [PubMed] [Google Scholar]
- 5.Krasnoff SB, Sommers CH, Moon Y-S, Donzelli BGG, Vandenberg JD, Churchill ACL, Gibson DM. J. Agric. Food Chem. 2006;54:7083–7088. doi: 10.1021/jf061405r. [DOI] [PubMed] [Google Scholar]
- 6.Moon YS, Donzelli BGG, Krasnoff SB, McLane H, Griggs MH, Cooke P, Vandenberg JD, Gibson DM, Churchill ACL. Appl. Environ. Microbiol. 2008;74:4366–4380. doi: 10.1128/AEM.00285-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donzelli BGG, Krasnoff SB, Churchill ACL, Vandenberg JD, Gibson DM. Curr. Genet. 2010;56:156–162. doi: 10.1007/s00294-010-0288-0. [DOI] [PubMed] [Google Scholar]
- 8.Faria M, Hajek AE, Wraight SP. Biol. Control. 2009;51:346–354. [Google Scholar]
- 9.Jegorov A, Havlicek V, Sedmera P. J. Mass Spectrom. 1998;33:274–280. doi: 10.1002/(SICI)1096-9888(199803)33:3<274::AID-JMS630>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 10.CCDC 840273 contains the crystallographic data for metacridamide A (1). These data can be obtained, free of charge, from The Cambridge Crystallographic Data Centre via http://www.ccdc.cam.ac.uk.
- 11.Schuemann J, Hertweck C. In: The Mycota XV: Physiology and Genetics. Anke T, Weber D, editors. Vol. XV. Berlin: Springer-Verlag; 2009. pp. 331–351. [Google Scholar]
- 12.Strieker M, Tanovic A, Marahiel MA. Curr. Opin. Struct. Biol. 2010;20:234–240. doi: 10.1016/j.sbi.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 13.Song Z, Cox RJ, Lazarus CM, Simpson TJ. ChemBioChem. 2004;5:1196–1203. doi: 10.1002/cbic.200400138. [DOI] [PubMed] [Google Scholar]
- 14.Eley KL, Halo LM, Song Z, Powles H, Cox RJ, Bailey AM, Lazarus CM, Simpson TJ. ChemBioChem. 2007;8:289–297. doi: 10.1002/cbic.200600398. [DOI] [PubMed] [Google Scholar]
- 15.Isaka M, Chinthanom P, Supothina S, Tobwor P, Hywel-Jones NL. J. Nat. Prod. 2010;12:279–289. doi: 10.1021/np100492j. [DOI] [PubMed] [Google Scholar]
- 16.Gao Q, Jin K, Ying SH, Zhang Y, Xiao G, Shang Y, Duan Z, Hu X, Xie XQ, Zhou G. PLoS Genet. 2011;7:879–920. doi: 10.1371/journal.pgen.1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacquot D, Poeschke O, Burger C. WO/2009/146,772. International Patent. 2009
- 18.Pittayakhajonwut P, Usuwan A, Intaraudom C, Khoyaiklang P, Supothina S. Tetrahedron. 2009;65:6069–6073. [Google Scholar]
- 19.Stratmann K, Burgoyne DL, Moore RE, Patterson GML, Smith CD. J. Org. Chem. 1994;59:7219–7226. [Google Scholar]
- 20.Fex T. Tetrahedron Lett. 1981;22:2703–2706. [Google Scholar]
- 21.Cantrell CL, Gustafson KR, Cecere MR, Pannell LK, Boyd MR. J. Am. Chem. Soc. 2000;122:8825–8829. [Google Scholar]
- 22.Molnár I, Gibson DM, Krasnoff SB. Nat. Prod. Rep. 2010;27:1241–1275. doi: 10.1039/c001459c. [DOI] [PubMed] [Google Scholar]
- 23.Szebenyi DME, Arvai A, Ealick S, LaIuppa JM, Nielsen C. J. Synchrotron Radiat. 1997;4:128–135. doi: 10.1107/S0909049597000204. [DOI] [PubMed] [Google Scholar]
- 24.Thorne RE, Stum Z, Kmetko J, O'Neill K, Gillilan R. J. Appl. Crystallogr. 2003;36:1455–1460. [Google Scholar]
- 25.Kabsch W. J. Appl. Crystallogr. 1993;26:795–800. [Google Scholar]
- 26.Kabsch W. Acta Crystallogr. Sect. D. Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheldrick GM. Acta Crystallogr. Sect. A: Found. Crystallogr. 2008;A64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 28.Krasnoff SB, Lobkovsky EB, Wach MJ, Loria R, Gibson DM. J. Agric. Food Chem. 2005;53:9446–9451. doi: 10.1021/jf051614w. [DOI] [PubMed] [Google Scholar]
- 29.Krasnoff SB, Gibson DM, Belofsky GN, Gloer KB, Gloer JB. J. Nat. Prod. 1996;59:485–489. [Google Scholar]
- 30.Roehm NW, Rodgers GH, Hatfield SM, Glasebrook AL. J. Immunol. Methods. 1991;142:257–265. doi: 10.1016/0022-1759(91)90114-u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.