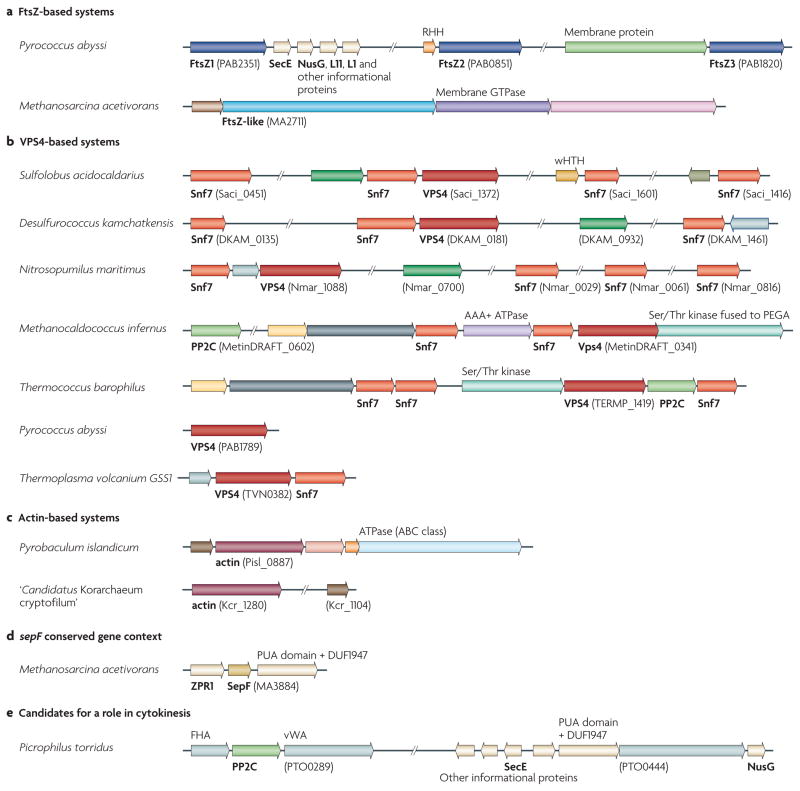
Figure 1. representative gene neighbourhoods of known and predicted membrane remodelling and cell division systems in the Archaea.
Homologous genes are the same colour; genes are shown to scale (with the exception of various informational genes shown in cream). Specific domains predicted to be encoded in the genes are listed above the gene, and predicted proteins (or the names of their eukaryotic homologues) are given below the gene in bold. The locus identifier is indicated for one gene in each putative operon (bracketed text). Representative gene neighbourhoods are shown from selected species. Complete information is available in Supplementary information S4,S5 (figure, table). a | The gene organization of the FtsZ-encoding gene and FtsZ-like protein-encoding genes of two representative organisms with an FtsZ or FtsZ-like division machinery. b | Representative organizations of the genes encoding two components of ESCRT (endosomal sorting complex required for transport), vacuolar protein sorting 4 (VPS4) and Snf7. c | An overview of the genetic organization of the genes encoding actin-like proteins. d | The genetic neighbourhood of the gene encoding the SepF homologue in Methanosarcina acetivorans. e | The genetic neighbourhood of the genes encoding proteins that are potentially involved in cytokinesis in Picrophilus torridus, which lacks the FtsZ, ESCRT and actin-based systems. AAA+ ATPase, ATPase associated with various cellular activities; FHA, forkhead-associated domain; l1, ribososmal protein l1; l11, ribosomal protein l11; PP2C, protein phosphatase 2C; RHH, ribbon–helix–helix domain (a predicted transcriptional regulator); vWA, von Willebrand factor type A domain; wHTH, winged helix–turn–helix domain; ZPR1, zinc finger protein 1.