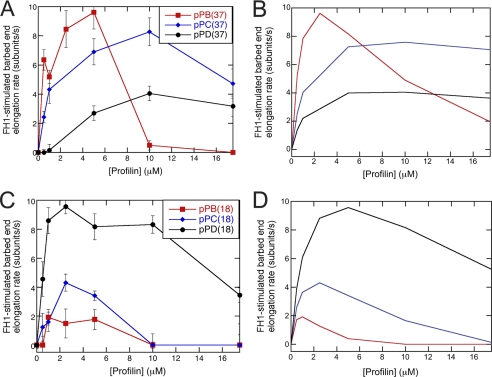
FIGURE 4.
Effect of sequence variations on FH1-stimulated polymerization. Conditions: 1.5 μm actin (33% Oregon Green) in microscopy buffer with varying concentrations of profilin. Data were collected by TIRFM. A, experimental and B, simulated dependence of FH1-stimulated barbed-end elongation on profilin concentration for variants of Bni1p with a single polyproline track (pPB, pPC, or pPD) located 37 residues (pPB position) from the FH2 domain. The error bars are S.E. C, experimental and D, simulated dependence of FH1-stimulated barbed-end elongation on profilin concentration for variants of Bni1p with a single polyproline track (pPB, pPC, or pPD) located 18 residues (pPD position) from the FH2 domain. The sequences inserted at these positions are the polyproline track sequences from sites pPB (red squares), pPC (blue diamonds), or pPD (black circles). Dissociation equilibrium constants for profilin:polyproline interactions are 20 μm for pPB, 600 μm for pPC, and 1200 μm for pPD. The loop closure rate for the profilin-bound FH1 domain is 10,000 s−1 at the pPB position and 50,000 s−1 at the pPD position. To account for a loss of FH1 flexibility upon insertion of the pPB sequence at the pPD position, the loop closure rate for free FH1 domain is 5,000 s−1 for pPB18FH2.