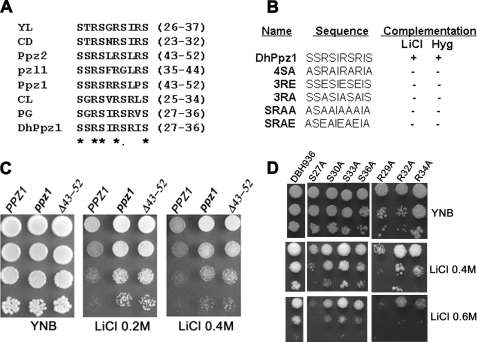
FIGURE 6.
Mutational analysis serine/arginine-rich motif in DhPpz1p and Ppz1p. A, sequence alignment of the serine/arginine-rich motif present in different Ppz1p orthologs from different species is shown: YL, Yarrowia lipolytica XP_504610.2; CD, Candida dubliniensis XP_002422157.1); Pzl1, Neurospora crassa AF071752; CL, Clavispora lusitaniae XP_002619471; PG, Pichia guilliermondii EDK41476.2; Ppz1, S. cerevisiae NP_013696.1; Ppz2, S. cerevisiae NP_010724.1; DhPpz1, D. hansenii XP_459586.2. Conserved residues are shown by asterisks. B, phenotypic complementation of wild type DhPpz1p and mutants carrying point mutations in serine/arginine-rich motif along with the sequence of respective mutated region are shown. + indicates complementation, and − indicates not complementation. C, dilution spotting of S. cerevisiae strains expressing wild type Ppz1p or its mutants on SD plate containing LiCl is shown. PPZ1 (parental strain BY4742/pRS423), ppz1 mutant (Y10557/pRS423), and Δ43–52 (Y10557/PPZ1-Δ43–52) were grown overnight on SD without histidine before serial dilution. Representative data of three independent experiments are shown. D, effect of mutation in individual serine and arginine residues of Ser/Arg motif on salt tolerance exhibited by dhppz1 mutant is shown. 10-Fold serial dilution of the DBH936 strain expressing different point mutations was spotted on SD plates containing 0.4 and 0.6 m LiCl. Plates were incubated at 28 ºC for 3–4 days before being photographed. A culture of DBH936 harboring empty vector pDA1 was used as the control. Representative data of two independent experiments are shown.