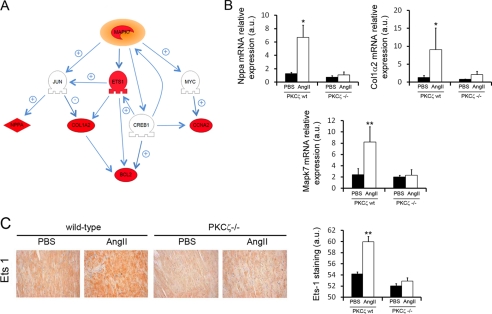
FIGURE 5.
Alterations in gene expression related to the activation of the ERK5 signaling pathway differ in wild-type and PKCζ-deficient mice. A, microarray data were analyzed using Pathway Studio to identify changes in the expression of genes functionally related to ERK5. Several genes that are part of the ERK5 signaling network were found up-regulated (red) in angiotensin II (AngII)-treated mice compared with PBS-treated mice or angiotensin II-treated PKCζ-deficient animals. Genes whose expression does not change are shown in gray. The interaction map shows reported positive (+) and negative (−) relationships between related proteins. Mapk7 (ERK5), jun (c-jun), Ets1 (ETS1), myc (c-Myc), Nppa (ANP, atrial natriuretic factor), Nppb (BNP, brain type natriuretic peptide), Col1α2 (collagen type I, α2), Ccna2 (cyclin A2), Creb1 (CREB, cAMP-responsive element binding protein 1), Bcl2 (BCL2). B, quantitative RT-PCR analysis in heart tissue showing enhanced mRNA expression of Nppa, Col1α2 and Mapk7 in angiotensin II-treated wild-type mice compared with angiotensin II-treated PKCζ−/− mice and vehicle-treated animals. Data are mean ± S.E. of three independent experiments. *, p < 0.05; **, p < 0.01, two-tailed t test when compared with untreated animals. C, immunohistochemical analysis with a specific antibody shows the overexpression of ETS1 in heart sections from angiotensin II-treated wild-type but not in control and PKCζ-deficient animals. Staining intensity was quantified and plotted as described in the legend to Fig. 3. Data are mean ± S.E. of three independent experiments. **, p < 0.01, two-tailed t test when compared with untreated animals. a.u., arbitrary units.