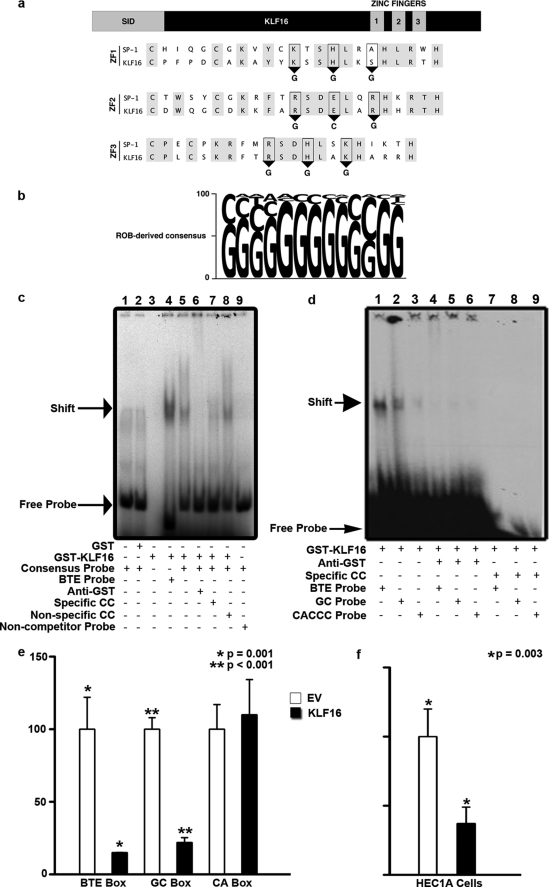
FIGURE 1.
KLF16 DNA binding domain displayed selectivity for distinct cis-regulatory elements. a, sequence alignment of the zinc fingers of SP1 and KLF16 revealing critical conserved residues (gray). Predicted KLF16 binding to GGG, GCG, and GGG bases via zinc fingers 1–3, respectively (ZF 1, 2, 3), is shown. b, ROB assay for KLF16 in uterine cells. 32P-Labeled ROB oligonucleotides were incubated with GST-KLF16 and separated by nondenaturing gel electrophoresis. The consensus sequence derived from 45 sequenced oligonucleotides after seven rounds of ROB is shown. The font size of each nucleotide corresponds to frequency of occurrence. c, EMSA using either 32P-labeled consensus KLF16-ROB (lanes 1, 2 and 5–8), BTE (lane 4), or noncompetitor probe (lane 9) with either 1 μg of GST protein (lane 2), GST-KLF16-zinc finger (ZF) (lanes 3–9), or probe alone (lane 1). Where indicated, the following were also added: 1 μg of anti-GST (lane 6), 500 m excess of unlabeled KLF16-ROB consensus probe (cold competitor, lane 7), or 500 m excess unlabeled noncompetitor probe (lane 8). Specific complexes formed between GST-KLF16 and either labeled BTE or KLF16-ROB probe (lanes 5 and 8) are indicated by an arrow. Anti-GST disrupted the GST-KLF16-ZF/consensus probe complex (lane 6). Addition of excess unlabeled consensus probe competed for the binding (lane 7), whereas an unrelated noncompetitor probe did not (lane 8). GST-KLF16-ZF did not shift the labeled noncompetitor probe (lane 9). Lane 1 contained only labeled KLF16-ROB consensus probe and no protein, whereas lane 3 contained 1 μg of GST-KLF16-ZF fusion protein alone. d, EMSA comparing GST-KLF16-ZF (1 μg per lane; lanes 1–9) protein binding to either 32P-labeled CYP1A1 BTE (lanes 1, 4, and 7), KLF consensus element (GC box) (lanes 2, 5, and 8), and CA box (lanes 3, 6, and 9). GST-KLF16-ZF protein (lanes 1–3) specifically bound all three KLF-binding elements (lanes 1–3, arrow indicates the shift), the BTE box with highest affinity (lane 1), robust binding to the GC box (lane 2), and the CA box with least affinity (lane 3). KLF16 binding to all elements was specifically disrupted by 1 μg of anti-GST (lanes 4–6: lane 4, BTE; lane 5, GC; lane 6, CA). Binding was lost on addition of 500 m fold excess unlabeled specific cold competitor (lanes 7–9: lane 7, BTE; lane 8, GC; lane 9, CA). e, uterine cells were cotransfected with 7.5 μg of FLAG-KLF16 or corresponding empty vector and either a 6×-BTE-, 6×-GC-, or 6×-CA-luciferase reporter vector (2.5 μg). Luciferase levels normalized to total protein levels and binding showed significant repression compared with empty vector, when KLF16 was cotransfected with either the 6×BTE (*, p = 0.001) or 6×GC-luciferase reporter (**, p < 0.001). There was no repression of 6×-CA-luciferase (f). HEC1A uterine cells were cotransfected for 48 h with either 7.5 μg of FLAG-KLF16 or corresponding parent pCMV/FLAG vector and 2.5 μg of luciferase reporter containing 6×-tandem BTEs. Luciferase activity normalized to protein concentrations show that compared with EV, KLF16 decreased luciferase expression by 63% (*, p = 0.003).