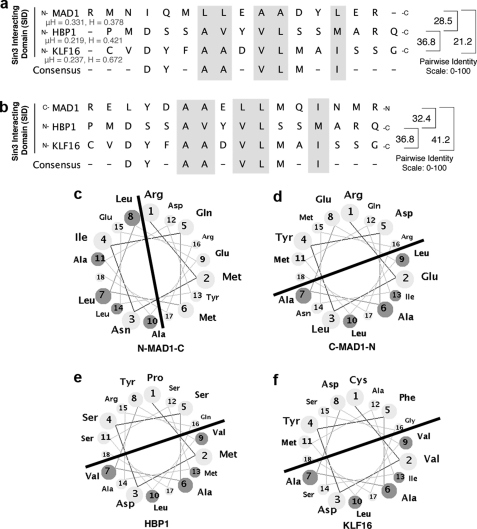
FIGURE 5.
Diagrammatic representation analyses of the MAD1, HBP1, and KLF16 SIDs, multiple SID sequence alignment. Forward (N to C terminus) (a) and reverse (C to N terminus) (b) SID alignments suggest that peptide polarity influences the interaction between the KLF16 SID with the Sin3a PAH2 domains, as the maximal similarity score derived by from pairwise alignments is obtained when the SID of KLF16 and HBP1 adopt the reversed orientation to MAD1. Overall, the high similarity among these peptides in hydrophobicity (H) and hydrophobic moment (μH) along with our previously published circular dichroism studies strongly support that the modeling of KLF16 SID as an amphipathic α-helix is appropriate (18). Comparative helical representation of the MAD1 (c and d), HBP1 (e), and KLF16 (f) SID; similar to MAD1 and HBP1 SID, KLF16 SID is predicted to form an amphipathic α-helix. The line separates the half of the helix containing the grouping of hydrophobic amino acid likely to interact with the Sin3a PHA2 domain. Noteworthy, as with the linear peptide alignment, when these helical wheels are built from the N to C orientation, the hydrophobic part of these helices adopt different orientations, suggesting that the mechanism used by these SID to interact with PAH2 complex are similar in HBP1 and KLF16 but different from MAD1. The main SID residues responsible for these interactions are either identical or conservative substitutions. Based on the sequence analyses of both proteins, these residues within the helix are critical for interacting with PAH2 and form the consensus DXXA(A/V)ϕVLXXIM. Helices were built using the Helical Wheel Applet from Virginia University.