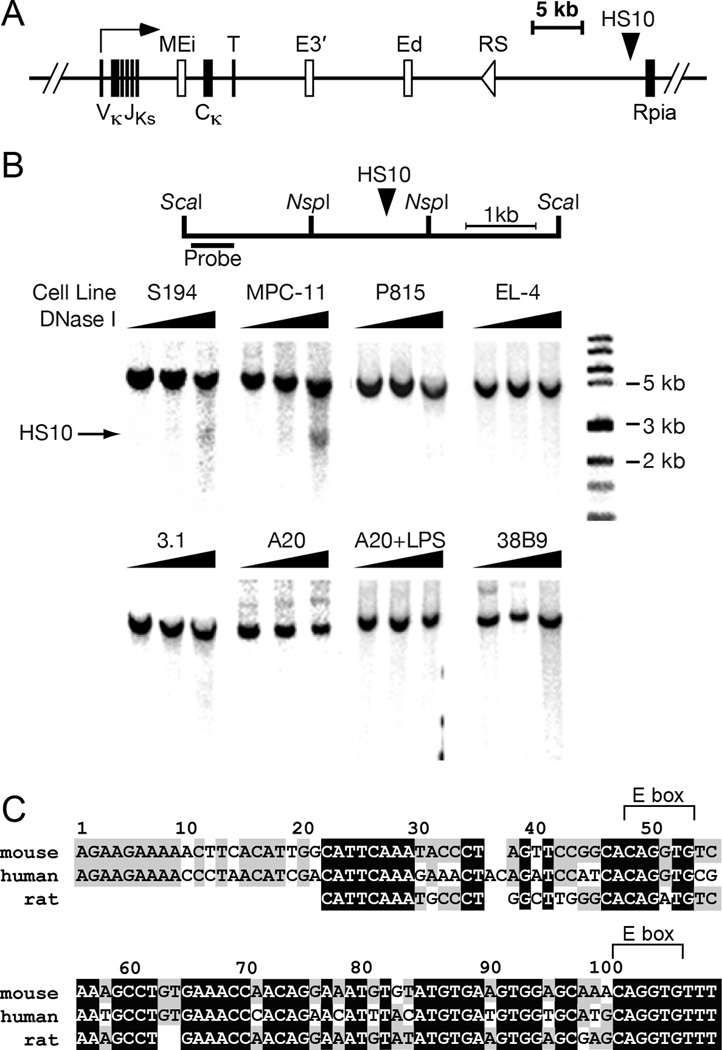
FIGURE 1.
Mapping and sequence analysis of DNase I hypersensitive site HS10. A, Schematic diagram depicting a rearranged Igκ locus. The coordinates of Ei, E3′, Ed, and HS10 in the NCDI37/mm9 mouse chromosome 6 sequence are respectively 70,675,570 to 70,676,084, 70,685,250 to 70,686,058, 70,693,704 to 70,694,944, and 70,713,758 to 70,715,084. B, upper: Schematic diagram of the positions of ScaI and NspI restriction sites, HS10 and the hybridization probe. lower: HS10 was mapped by indirect end-labeling after DNase I digestion of the various indicated permeabilized cell lines and ScaI digestion of the resulting purified DNA followed by Southern blotting. The solid arrow indicates the position of HS10, which is plasmacytoma-cell-specific. Cell lines correspond to plasmacytomas S194 and MPC-11 (25, 26), mastocytoma P815 (27), pro-B 38B9 (28), pre-B 3-1 (28), mature B A20 (29), and mature T EL-4 cells (30). C, HS10 sequences are conserved between mouse, human and rat. Blank spaces indicate no match, gray and black boxes indicate two or three matches between species, respectively. Two conserved E-boxes (CANNTG) are shown above the sequences.