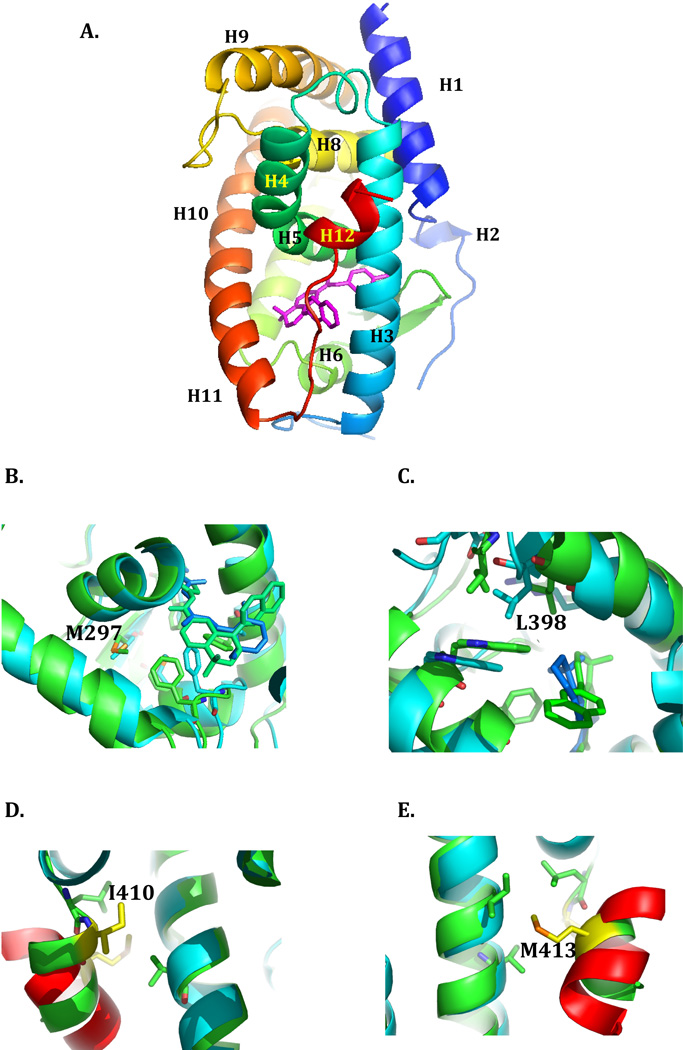
Figure 1. The ligand binding domain of RARα.
The structure of the ligand binding domain of RARα in its antagonist bound form (PDB ID:1DKF) is depicted in ribbon representation, with the N- to C-terminus in rainbow coloring. The individual helices are labeled, and the positions of mutations described in the text indicated. The antagonist ligand BMS600 is stick rendered in magenta. Figure 1 was generated using PyMOL 68. The sources of the mutants (summarized in 35) are as follows: M297L, referred to as APL Case 2, was first described in 45; L398P is referred to in the literature as NB4-R4 44; I410T is referred to as NB4-A1 49; M413T is referred to APL Case 9 46. The LBDs of RARα in the antagonist bound state (PDB:1DKF) and RARβ in the agonist bound state (PDB:1XDK) were superimposed using the MagicFit option of Swiss PDB Viewer. The RARα (antagonist bound) structure is colored green, and the RARβ (agonist bound) is colored cyan. The fragment of the TRAP220 co-activator bound to the RARβ is colored red. A, the region around Met297. B, the region around Leu398. C, the region around Ile410. D, the region around Met 413.