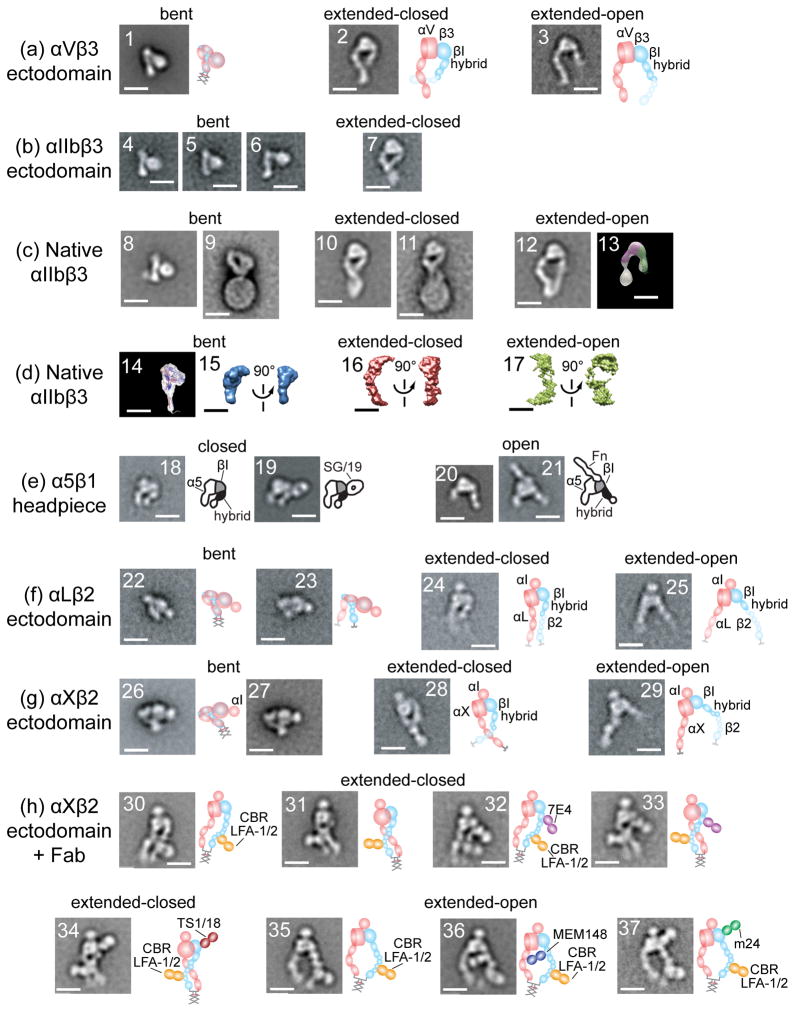
Figure 2.
Structure of integrins from electron microscopy, electron tomography, and neutron or X-ray scattering in solution reveal three conformational states. Schematics are shown to right. All scale bars = 10 nm. With permission from cited references. Panels show representative class averages of negatively-stained integrins unless otherwise noted. (a) αVβ3 ectodomain with a C-terminal coiled-coil clasp (1) or unclasped with Mn2+ (2) or RGD (3) [13]. (b) αIIbβ3 ectodomain, clasped (4) or unclasped (5-7) [3]. (c) αIIbβ3 purified from platelets in detergent (8, 10, 12) [23], (13) [16] and embedded in lipoprotein nanodiscs (9, 11) [27]. With no additions (8-9), Mn2+ (10), talin head domain (11), and RGD peptide or RGD mimetics (12-13). Negative stain electron tomography class average (13). (d) Three-dimensional molecular envelopes of purified, detergent-soluble native αIIbβ3 in solution determined by small-angle neutron scattering (14) [21] or X-ray scattering (15-17) [23] with no additions (14-15), Mn2+ (16), or Mn2+ and RGD mimetic (17). (e) α5β1 headpiece [14,15] alone (18), + allosteric inhibitory SG/19 Fab (19), + RGD (20), or + fibronectin domain 7-10 fragment (21). (f) LFA-1 ectodomain [19] clasped (22) or unclasped (23-25). (g) αXβ2 [17,19] clasped from different publications (26-27) and unclasped (28-29). (h) αXβ2 [17,19] clasped with extension and activation-promoting CBR LFA-1/2 Fab (30, 31, 35); and with CBR LFA-1/2 Fab and allosteric inhibitory Fab 7E4 (32-33) and TS1/18 (34) or allosteric activating Fab MEM148 (36) or m24 (37).