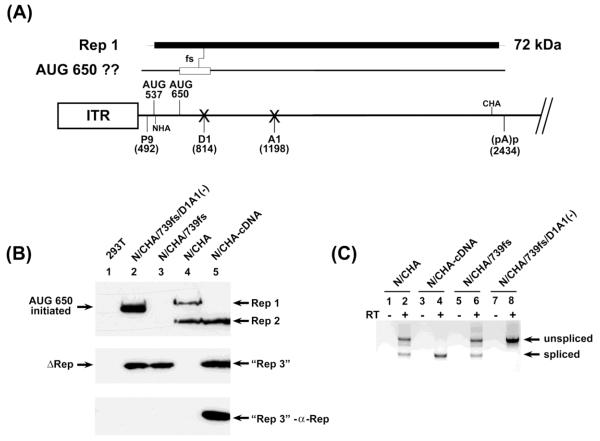
Figure 3. Competent splice sites prevented 650 AUG usage in unspliced RNA.
(A): The map shows potential products from unspliced RNAs and the frame shift mutant described. The central line depicts the genome as described in Fig. 1, with “Xs” at the donor and acceptor site to indicate their mutation, and positions of both the N-terminal and C-terminal HA tags, at nts 540 and 2241, respectively. Above the line is depicted the coding region for Rep1 and the site of the frame shift described, and the potential protein generated by this mutant if the 650 AUG is used. (B): The left panel is an immunoblot of protein extracts from 293T cells either alone, or transfected with the constructs indicated and as described in the text. The upper two panels are reacted with anti-HA antibody. All of these constructs contained both the N- and C-terminal HA tag in ORF 1 at nt 540 and nt 2241, respectively. The wild-type RepCap clone as well as the cDNA-like clone, both also doubly tagged (N/CHA and N/CHA-cDNA, respectively), were included as controls. Rep1, Rep 2 and the “Rep3-like” proteins, reacting with anti-HA antibody, are indicated. The bottom-most panel shows that the “Rep3-like” protein (lane 4), but not the similarly migrating Δ-Rep proteins (lane 2 and 3), is authentic Rep3, as it also reacts with an anti-Rep antibody raised to the unique portion of Rep3 shown in Figure 1 and as described in the text and Materials and Methods. (C): The panel to the lower right is an RT-PCR of RNAs extracted from the same transfection indicating splicing levels using primers spanning the splice junction.