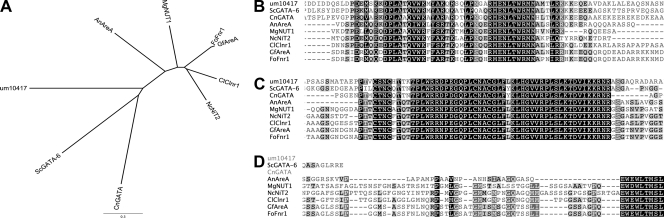
Fig 1.
Sequence analysis of U. maydis GATA transcription factor Um10417. (A) Phylogenetic tree of characterized Nit2/AreA transcription factors from the ascomycetes Aspergillus nidulans (AnAreA), Colletotrichum lindemuthianum (ClClnr1), Fusarium oxysporum (FoFnr1), Gibberella fujikuroi (GfAreA), and Neurospora crassa (NcNit2), as well as homologous proteins from the basidiomycetes Schizophyllum commune (ScGata-6) and Cryptococcus neoformans (CnGat1). (B to D) Protein alignment showing the DUF1752 motif conserved among functional Nit2/AreA proteins (B), the Zn finger motif (C), and the C-terminal motif (D) conserved only in ascomycete Nit2/AreA proteins. Multiple sequence alignment was executed using the MUSCLE algorithm (20).