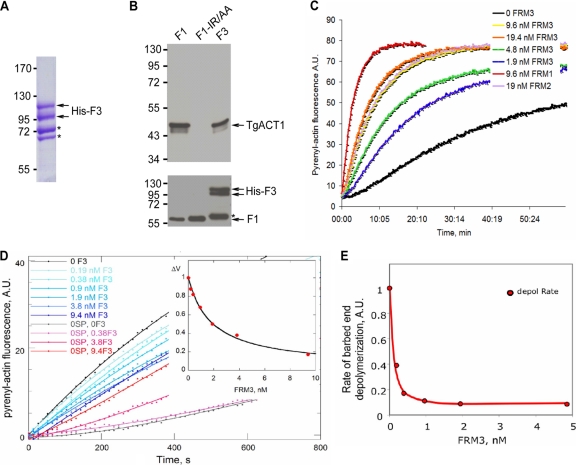
Fig 2.
The recombinant TgFRM3 FH2 domain (F3) binds to TgACT1 and polymerizes rabbit actin. (A) Separation of the His-tagged F3 domain expressed in E. coli (BL21) and purified on a nickel column. The F3 protein was loaded on an 8% SDS-polyacrylamide gel, followed by Coomassie blue staining. Asterisks indicate bacterial histidine-rich proteins or chaperone binding to nickel beads, which are eluted with the F3 domain under native conditions. (B) Nickel affinity pulldown assay, measuring the abilities of the TgFRM1 FH2 domain (F1), the F1 actin binding domain mutant (F1-IR/AA), and the F3 domain fused to His to bind to TgACT1. The amounts of TgACT1 and FH2 domains were determined by Western blot analysis using antiactin and anti-His antibodies. The asterisk represents F3 domain degradation. (C) F3 nucleates actin assembly in vitro. Spontaneous actin assembly was recorded in the presence of F3 at the indicated concentrations. (D) F3 inhibits barbed-end growth of actin filaments. The initial rate of barbed end growth was measured at different concentrations of F3 in the presence (blue curves) and in the absence (red curves) of 0.11 nM spectrin-actin (SP) seeds. The normalized difference between the rates measured with and without seeds is plotted versus the concentration of F3 (inset). The calculated curve fits the data with an affinity of 0.55 nM−1 for binding of F3 to barbed ends, leading to 85% ± 5% inhibition of barbed-end growth. (E) F3 caps barbed ends and inhibits barbed-end depolymerization with high affinity. The rate of dilution-induced depolymerization was about 90% inhibited by F3 binding to the barbed ends, with an affinity of 10 nM−1.