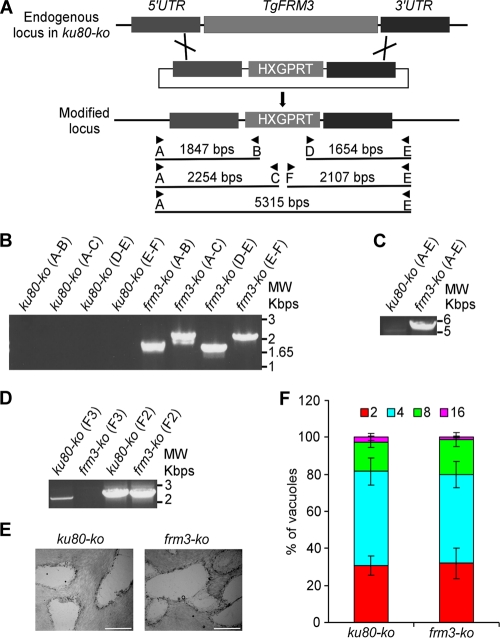
Fig 4.
Generation of a TgFRM3 knockout. (A) Schematic representation of the strategy used to replace the TgFRM3 open reading frame with the HXGPRT resistance cassette. The knockout plasmid for TgFRM3 contains 1,497 bp of the 5′ untranslated region (UTR) of TgFRM3 (dark gray), the HXGPRT gene (light gray), and 1,498 bp of the 3′ untranslated region (UTR) of TgFRM3 (dark gray). Black arrows represent the primers used for PCR analysis, and the lengths of the PCR products generated are indicated. (B and C) PCR analysis performed on the frm3-ko strain, showing that double homologous recombination occurred. Genomic DNA from ku80-ko parasites was used as a control. (D) Endogenous-locus PCR analysis of ku80-ko and frm3-ko parasites, which shows that TgFRM3 open reading frame mRNA was undetectable in frm3-ko parasites. The TgFRM2 FH2 domain (F2) served as a loading control. (E) Plaque assay performed on an HFF monolayer infected with ku80-ko or frm3-ko parasites. After 7 days, the HFF were stained with Giemsa stain. The scale bar represents 1 mm. (F) Intracellular growth of ku80-ko and frm3-ko strains cultivated in parallel for 48 h. Freshly released parasites from both strains invade new HFF cells. Numbers of parasites per vacuole were counted at 24 h after inoculation. The percentage of vacuoles containing a given number of parasites is represented on the y axis. Values are means ± standard deviations (SD) for three independent experiments.