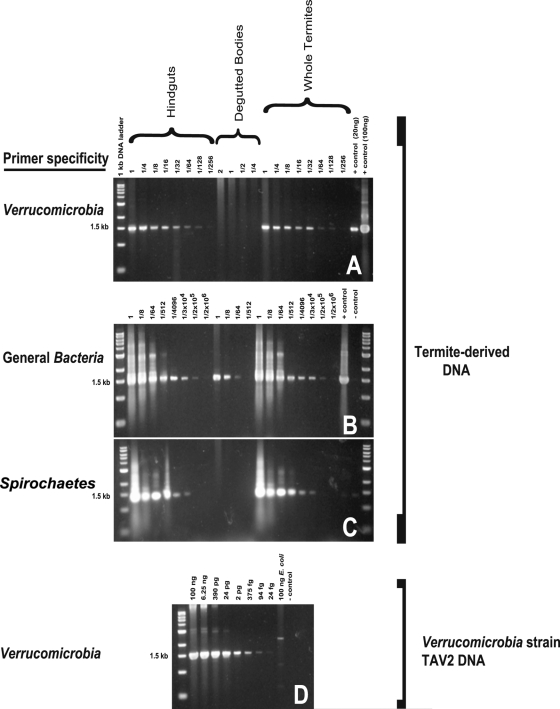
Fig 1.
(A to C) Dilution-to-extinction PCR of DNA from termite hindguts, degutted termite bodies, and whole termites amplified with Verrucomicrobia (A)-, general Bacteria (B)-, and Spirochaetes (C)-specific primers. Numbers above each lane represent the amount of DNA added on a “per-termite-equivalent” basis. (D) Purified DNA from Verrucomicrobia strain TAV2 was serially diluted and amplified as a control. Assuming that the average nucleotide has a molecular mass of 324 g/mol, one 5.2-Mb TAV2 genome with one 16S rRNA gene copy is slightly over five femtograms. A PCR product was detected in termite gut DNA diluted to the equivalent of 1/256 of a gut (A). A PCR product was detected when control TAV2 DNA was diluted to 24 fg, approximating 4.8 cells (D). Thus, we can estimate that there are 4.8 verrucomicrobia cells per 1/256 of a gut, yielding a total of 1.2 × 103 verrucomicrobial cells per termite gut.