Abstract
Common laboratory strains of Bacillus subtilis encode two glutamate dehydrogenases: the enzymatically active protein RocG and the cryptic enzyme GudB that is inactive due to a duplication of three amino acids in its active center. The inactivation of the rocG gene results in poor growth of the bacteria on complex media due to the accumulation of toxic intermediates. Therefore, rocG mutants readily acquire suppressor mutations that decryptify the gudB gene. This decryptification occurs by a precise deletion of one part of the 9-bp direct repeat that causes the amino acid duplication. This mutation occurs at the extremely high frequency of 10−4. Mutations affecting the integrity of the direct repeat result in a strong reduction of the mutation frequency; however, the actual sequence of the repeat is not essential. The mutation frequency of gudB was not affected by the position of the gene on the chromosome. When the direct repeat was placed in the completely different context of an artificial promoter, the precise deletion of one part of the repeat was also observed, but the mutation frequency was reduced by 3 orders of magnitude. Thus, transcription of the gudB gene seems to be essential for the high frequency of the appearance of the gudB1 mutation. This idea is supported by the finding that the transcription-repair coupling factor Mfd is required for the decryptification of gudB. The Mfd-mediated coupling of transcription to mutagenesis might be a built-in precaution that facilitates the accumulation of mutations preferentially in transcribed genes.
INTRODUCTION
As the central amino group donor for nearly all biosynthetic pathways in any living cell, glutamate plays a key role in the biochemistry and physiology of all organisms (15). Investigations with Escherichia coli demonstrate that glutamate is by far the most abundant metabolite in these bacteria, accounting for ca. 40% of the internal metabolite pool (60). Moreover, glutamate is one of the most highly embedded metabolites. In the Gram-positive soil bacterium Bacillus subtilis, at least 37 reactions make use of this amino acid (42).
In B. subtilis, glutamate is exclusively synthesized from 2-oxoglutarate and glutamine by the activity of glutamate synthase in the absence of exogenous glutamate or other sources of glutamate. 2-Oxoglutarate is replenished in the citric acid cycle, whereas glutamine can be synthesized with ammonium as the nitrogen source and one of the two molecules of glutamate that are generated by glutamate synthase as the acceptor. Glutamate does also serve as a precursor for proline biosynthesis and, under conditions of osmotic stress, molar concentrations of proline have to be produced (28). Thus, it is not surprising that glutamate synthesis has to be a highly efficient process and, indeed, interactions between enzymes of the branch of the citric acid cycle that generates 2-oxoglutarate and glutamate synthase have been reported (39). Glutamate can also serve as source of carbon and nitrogen. Its utilization is initiated by an oxidative deamination catalyzed by the glutamate dehydrogenase. The expression of the genes encoding glutamate biosynthetic and catabolic enzymes is subject to complex control mechanisms that allow the adjustment of the intracellular glutamate concentration to the actual requirement (6, 7, 16, 44, 51).
B. subtilis encodes two glutamate dehydrogenases, GudB and RocG (5). However, the gudB gene experienced an inactivating mutation during domestication, resulting in an inactive pseudogene in the laboratory strain B. subtilis 168. In contrast, the gudB gene encodes an active enzyme in wild isolates and in nondomesticated strains such as NCIB3610 (61). The inactivation of gudB is caused by a duplication of nine base pairs of the coding sequence resulting in a duplication of three amino acids in the active center of the protein. The glutamate dehydrogenase RocG catalyzes the final step of the catabolic pathway for arginine, ornithine and citrulline. Accordingly, its expression is strongly induced in the presence of arginine (5). Interestingly, the glutamate dehydrogenases are not only required for glutamate utilization, but they are also involved in the control of glutamate biosynthesis: in the presence of glutamate they inhibit the transcription activator GltC that is necessary for the expression of the glutamate synthase operon, gltAB (8, 16, 18, 25). In the active state, the two glutamate dehydrogenases are very similar to each other, both at the level of the amino acid sequence and also concerning their structures. In contrast, the inactive GudB protein seems to misfold and is subject to rapid degradation (23, 25).
The importance of glutamate for the cellular physiology is underlined by the observation that any mutation that disturbs the glutamate homoeostasis results in the accumulation of suppressing mutations. This is true for both E. coli and B. subtilis (19, 59). In the laboratory strain of B. subtilis, the inactivation of the rocG gene encoding the only active glutamate dehydrogenase results in the appearance of mutants with an active GudB enzyme (these alleles are designated gudB1) (5). Moreover, rocG gudB double mutants easily acquire suppressive mutations affecting the glutamate synthase (19). The rocG gudB double mutants are unable to utilize glutamate as the single source of carbon. However, cultivation of such mutants in the presence of glutamate or its precursors such as arginine results in the selection of suppressor mutants that catabolize glutamate by a pathway that is not operative in wild-type bacteria. An analysis of one such mutant revealed constitutive expression of the aspartase pathway due to the inactivation of the repressor of the corresponding ansAB operon, AnsR (21).
The accumulation of mutations that restore growth of mutants or that allow faster growth is a common phenomenon in bacteria. Several studies suggest that mutations that overcome the specific limitation are preferentially acquired (3, 13); however, the underlying mechanisms have not yet been elucidated.
Mutations can be acquired during replication. Most of the errors are eliminated by DNA mismatch repair, including the MutSL system which contributes to genome stability (22, 40). Some errors can escape from repair and may be beneficial for the organism. Many bacteria, including B. subtilis, possess systems for the induction of mutations in the stationary phase (55). The emergence of these mutations is associated with transcription rather than with DNA replication and plays an important role in the generation of diversity in nondividing populations of B. subtilis. The process of transcription-coupled DNA repair is crucial for the accumulation of mutations in the stationary phase, and this involves the transcription repair coupling factor Mfd (2). The Mfd protein targets DNA lesions during transcription that provoked a roadblock of transcription. Subsequently, Mfd may displace the RNA polymerase and recruit the nucleotide excision repair system to resolve the lesion (12, 56). It was suggested that this process favors the acquisition of beneficial mutations of highly transcribed genes (45, 46).
We are interested in the mechanism by which the decryptification of the gudB gene occurs in rocG mutants. The gudB1 mutation appears during growth and requires a deletion of 9 bp. Therefore, gudB provides a unique system to study the emergence of mutations. Our results suggest that the decryptification of gudB requires the presence of a perfect direct repeat. Moreover, a part of this repeat is preferably deleted with a high frequency in the context of a transcribed gene, and this deletion requires the Mfd transcription repair coupling factor.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All B. subtilis strains used in the present study are derived from the laboratory wild-type strain 168. They are listed in Table 1. E. coli DH5α (48) was used for cloning experiments. B. subtilis was grown in SP medium, in LB medium, or in C minimal medium supplemented with tryptophan (at 50 mg liter−1) (58). CSE medium is C minimal medium supplemented with sodium succinate (6 g liter−1) and potassium glutamate (8 g liter−1). C-Glc is C minimal medium supplemented with glucose (1 g liter−1), and CS is supplemented with sodium succinate (6 g liter−1) (58). Additional sources of carbon and nitrogen were added as indicated. E. coli was grown in LB medium, and transformants were selected on plates containing ampicillin (100 μg/ml). LB, SP, and CS plates were prepared by the addition of 17 g of Bacto agar (Difco)/liter to LB, SP, or CS medium, respectively.
Table 1.
B. subtilis strains used in this study
| Strain | Genotype | Source or referencea |
|---|---|---|
| 168 | trpC2 | Laboratory collection |
| BG427 | trpC2 metB5 amyE sigB xin-1 attSP recU::cat | 20 |
| BP12 | trpC2 ΔgudB::aphA3 rocG::Tn10 spc amyE::(gudB1mut1 lacZ cat) | Spontaneous mutation of GP1179 on SP |
| BP13 | trpC2 ΔgudB::aphA3 rocG::Tn10 spc amyE::(gudB1mut3 lacZ cat) | Spontaneous mutation of GP1197 on SP |
| GP747 | trpC2 rocG::Tn10 spc | 17 |
| GP753 | trpC2 rocG::Tn10 spc gudB1 | Spontaneous mutation of GP747 on SP |
| GP754 | trpC2 rocG::cat amyE::(gltA-lacZ aphA3) | 16 |
| GP804 | trpC2 gudB1 amyE::(gltA-lacZ aphA3) | 19 |
| GP891 | trpC2 recU::cat | BG427→168 |
| GP892 | trpC2 rocG::spc recU::cat | BG427→GP747 |
| GP894 | trpC2 ΔsbcDC::aphA3 | See Materials and Methods |
| GP895 | trpC2 ΔrecJ::aphA3 | See Materials and Methods |
| GP896 | trpC2 rocG::spc ΔsbcDC::aphA3 | GP747→GP894 |
| GP897 | trpC2 rocG::spc ΔrecJ::aphA3 | GP747→GP895 |
| GP898 | trpC2 ΔexoA::aphA3 | See Materials and Methods |
| GP900 | trpC2 rocG::spc ΔexoA::aphA3 | GP747→GP898 |
| GP1101 | trpC2 amyE::(gudB-lacZ cat) | pGP651→168 |
| GP1102 | trpC2 gudB1 amyE::(gudB-lacZ cat) | pGP651→GP804 |
| GP1103 | trpC2 rocG::Tn10 spc recA::erm cat | IRN444→GP747 |
| GP1104 | trpC2 rocG::Tn10 spc amyE::(gudB-lacZ cat) | pGP651→GP747 |
| GP1105 | trpC2 rocG::Tn10 spc amyE::(gudB-lacZ cat) gudB1 | Spontaneous mutation of GP1104 on SP |
| GP1106 | trpC2 ΔaddAB::spc | HVS666→168 |
| GP1107 | trpC2 ΔaddAB::spc rocG::cat amyE::(gltA-lacZ aphA3) | GP1106→GP754 |
| GP1123 | trpC amyE::(alf-aphA3 lacZ cat) | pGP655→168 |
| GP1127 | trpC2 amyE::(alf1-aphA3 lacZ cat) | Spontaneous mutation of GP1123 on SP-Km |
| GP1160 | trpC2 ΔgudB::aphA3 | See Materials and Methods |
| GP1161 | trpC2 ΔgudB::aphA3 rocG::Tn10 spc | GP1160→GP747 |
| GP1163 | trpC2 ΔgudB::aphA3 rocG::Tn10 spc amyE::(gudB lacZ cat) | pGP900→GP1161 |
| GP1167 | trpC2 Δmfd::ermC | See Materials and Methods |
| GP1168 | trpC2 Δmfd::ermC amyE::(alf-aphA3 lacZ cat) | GP1167→GP1123 |
| GP1169 | trpC2 rocG::Tn10 spc Δmfd::ermC | GP1167→GP747 |
| GP1175 | trpC2 ΔuvrAB::Ermr | See Materials and Methods |
| GP1176 | trpC2 ΔuvrAB::ErmrrocG::Tn10 spc | GP1175→GP747 |
| GP1177 | trpC2 ΔgudB::aphA3 amyE::(gudBmut1 lacZ cat) | pGP1714→GP1160 |
| GP1178 | trpC2 ΔgudB::aphA3 amyE::(gudBmut2 lacZ cat) | pGP1715→GP1160 |
| GP1179 | trpC2 ΔgudB::aphA3 rocG::Tn10 spc amyE::(gudBmut1 lacZ cat) | GP747→GP1177 |
| GP1180 | trpC2 ΔgudB::aphA3 rocG::Tn10 spc amyE::(gudBmut2 lacZ cat) | GP747→GP1178 |
| GP1190 | trpC2 ΔmutSL::aphA3 | See Materials and Methods |
| GP1191 | trpC2 ΔmutSL::aphA3 rocG::Tn10 spc | GP747→GP1190 |
| GP1192 | trpC2 ΔmutSL::aphA3 ΔuvrAB::ErmrrocG::Tn10 spc | GP1176→GP1190 |
| GP1197 | trpC2 ΔgudB::aphA3 rocG::Tn10 spc amyE::(gudBmut3 lacZ cat) | pGP1721→GP1161 |
| GP1198 | trpC2 ΔgudB::aphA3 rocG::Tn10 spc amyE::(gudB1mut3 lacZ cat) | Spontaneous mutation of GP1197 on SP |
| GP1502 | trpC2 Δnfo::cat | See Materials and Methods |
| GP1503 | trpC2 ΔexoA::aphA3 Δnfo::cat | GP1502→GP898 |
| GP1504 | trpC2 ΔexoA::aphA3 Δnfo::cat rocG::spc | GP747→GP1503 |
| HVS666 | trpC2 ΔaddAB::spc | 14 |
| IRN444 | trpC2 recA::erm cat | 34 |
Arrows indicate construction by transformation.
DNA manipulation, transformation, and phenotypic analysis.
Transformation of E. coli and plasmid DNA extraction were performed according to standard procedures (48). Restriction enzymes, T4 DNA ligase, and DNA polymerases were used as recommended by the manufacturers. Phusion DNA polymerase was used for the PCR as recommended by the manufacturer. All primer sequences are provided as supplementary material (see Table S1 in the supplemental material). DNA sequences were determined using the dideoxy chain termination method (48). All plasmid inserts derived from PCR products were verified by DNA sequencing. Chromosomal DNA of B. subtilis was isolated as described previously (32).
E. coli transformants were selected on LB plates containing ampicillin (100 μg/ml). B. subtilis was transformed with plasmid or chromosomal DNA according to the two-step protocol described previously (32). Transformants were selected on SP plates containing kanamycin (10 μg/ml), chloramphenicol (5 μg/ml), spectinomycin (150 μg/ml), or erythromycin-lincomycin (2 and 25 μg/ml, respectively).
In B. subtilis, amylase activity was detected after growth on plates containing nutrient broth (7.5 g/liter), 17 g of Bacto agar (Difco)/liter, and 5 g of hydrolyzed starch (Connaught)/liter. Starch degradation was detected by sublimating iodine onto the plates.
Quantitative studies of lacZ expression in B. subtilis were performed as follows. Cells were grown in CSE medium supplemented with different carbon and nitrogen sources as indicated. The cells were harvested at an optical density at 600 nm (OD600) of 0.6 to 0.8 for cultures in CSE medium and an OD600 of 0.8 to 1.0 for cultures in CSE medium with sugar. β-Galactosidase specific activities were determined with cell extracts obtained by lysozyme treatment as described previously (32). One unit of β-galactosidase is defined as the amount of enzyme that produces 1 nmol of o-nitrophenol per min at 28°C.
Ectopic expression of gudB variants.
To express the gudB gene at an ectopic site, we used plasmid pAC5 (37). This plasmid allows integration of the cloned fragments into the amyE site of the B. subtilis chromosome. Briefly, the gudB gene was amplified with its natural promoter using the oligonucleotides ST1 and KG92 with the chromosomal DNA of B. subtilis 168 as the template. The PCR product was digested with EcoRI and BamHI and cloned into pAC5 linearized with the same enzymes. The resulting plasmid pGP900 was used to introduce the gudB allele into the chromosome.
The direct repeat of gudB present in pGP900 was subjected to site-directed mutagenesis by a modified PCR protocol, the combined chain reaction (9). Primers ST1 and KG92 were used as outer primers. The primers KG119, KG120, and KG133 were used to introduce point mutations into the gudB coding region. These primers were phosphorylated at their 5′ ends and allowed ligation of the nascent elongation product initiated from ST1. The resulting products carrying the mutations were cut with EcoI and BamHI and cloned into pAC5 digested with the same enzymes. The resulting plasmids were pGP1714 (G3T G9T), pGP1715 (G3T G9T G12T G18T), and pGP1721 (G12T G18T). The plasmids were linearized with PstI and used to transform B. subtilis (see Table 1).
Design and construction of a mutagenesis reporter system.
In order to analyze the occurrence of the deletion of the repeat in a nonrelated sequence context, we developed a reporter system that is based on a promoter that is only active upon deletion of one part of the gudB-derived direct repeat. This artificial alf promoter controls the expression of genes coding for a kanamycin-resistant determinant (aphA3) and E. coli β-galactosidase. To obtain the reporter strain, we first constructed plasmid pGP655 as follows. The promoterless aphA3 gene was amplified from pDG780 (24) by using the primer pair ST4 and ST9. These oligonucleotides attached restriction sites for EcoRI and BamHI (ST9) and for BglII (ST4) to the PCR product. The fragment was digested with EcoRI and BglII and cloned into the integration vector pAC6 (54), linearized with EcoRI and BamHI. The resulting plasmid pGP653 contained a promoterless aphA3-lacZ operon. The alf promoter fragment was obtained by hybridization of the complementary oligonucleotides ST7 and ST8. It was cloned between the EcoRI and BamHI sites of pGP653, resulting in plasmid pGP655.
Construction of mutant strains.
Deletion of the recJ, exoA, nfo, uvrAB, mutSL, sbcDC, gudB, and mfd genes was achieved by transformation with PCR products constructed using oligonucleotides (see Table S1 in the supplemental material) to amplify DNA fragments flanking the target genes and intervening antibiotic resistance cassettes (24), as described previously (57).
Construction of a gudB-lacZ fusion.
To determine the activity of the gudB promoter, a translational fusion of the gudB promoter to a promoterless lacZ gene encoding β-galactosidase was constructed as follows. A DNA fragment containing the gudB promoter region was generated by PCR using the primers ST1 and ST2, digested with BamHI and EcoRI, and cloned into the plasmid pAC5. The plasmid pAC5 contains a promoterless lacZ gene and allows the introduction of translational fusions into the amyE locus of B. subtilis (37). The resulting plasmid pGP651 was used to introduce the fusion into different B. subtilis mutants (see Table 1).
Determination of mutation frequencies.
The mutation frequencies were determined by the method of the median (33). Briefly, 11 cultures in CSE-Glc were inoculated to a density of 100 cells/ml with an overnight culture grown in the same medium. The cultures were incubated at 37°C to an OD600 of 2.0. For the analysis of culture titers appropriate dilutions of four cultures were plated on SP medium containing glucose to allow growth of the rocG mutant strains. To screen for gudB1 mutations, appropriate dilutions of each culture were plated on SP medium. After 24 h, the colonies showing the gudB1 phenotype (wild type-like colonies on SP plates) were counted. To be sure of the identity of the mutations, the gudB allele was sequenced for at least three independent suppressor mutants in each experiment. In every single case, the correct excision of one part of the repeat (i.e., the gudB1 mutation) was observed. For the determination of mutation frequencies of the alf promoter present in the strains GP1123 and GP1168, the bacteria were plated on SP medium containing kanamycin (60 μg/ml) and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 80 μg/ml).
Northern blot analysis.
Preparation of total RNA and Northern blot analysis were carried out as described previously (35). Digoxigenin (DIG) RNA probes were obtained by in vitro transcription with T7 RNA polymerase (Roche Diagnostics) using PCR-generated DNA fragments as templates. The primer pairs used to amplify DNA fragments specific for gudB and gapA are listed in Table S1 in the supplemental material. The reverse primers contained a T7 RNA polymerase recognition sequence. In vitro RNA labeling, hybridization, and signal detection were carried out according to the instructions of the manufacturer (DIG RNA labeling kit and detection chemicals; Roche Diagnostics). To determine the size of the gudB mRNA, we used the transcripts observed with a gapA probe as the standard. RNA stability was analyzed as described previously (38). Briefly, rifampin was added to logarithmically growing cultures (final concentration, 100 μg/ml), and samples were taken at the time points indicated. The quantification was performed using ImageJ software v1.42 (1).
Western blotting.
For Western blot analysis, proteins were separated by SDS–12.5% PAGE and transferred onto polyvinylidene difluoride membranes (Bio-Rad) by electroblotting. Rabbit anti-RocG (1:15,000) (17) served as the primary antibody. The antibodies were visualized by using anti-rabbit immunoglobulin G-alkaline phosphatase secondary antibodies (Promega) and the CDP-Star detection system (Roche Diagnostics), as described previously (16).
RESULTS
The gudB1 suppressor mutation appears at an extremely high frequency.
Previous studies revealed that the gudB gene readily acquired the gudB1 suppressor mutation if the rocG gene was inactivated (5, 17, 19). In order to describe this mutation event in a quantitative way, the frequency of the gudB reversion was determined. For this purpose, 11 independent cultures of the rocG mutant strain B. subtilis GP747 were inoculated with approximately 100 cells/ml to reduce the likelihood of very early mutants. The cultures were grown under nonselective conditions (in CSE medium supplemented with glucose) for 20 generations and plated on complex medium (SP medium), which is toxic for the rocG mutant but not for emerging rocG gudB1 suppressor strains. Suppressor mutants were recognized since they grew as solid colonies (like the wild-type strain B. subtilis 168), whereas the rocG mutant strain GP747 formed only very small opaque colonies on complex medium. To ascertain that the mutation had appeared during the cultivation and not as a result of selection on the plates, only suppressor mutants that were present after 24 h were taken into consideration. The mutation frequency was 10−4. To the best of our knowledge, such a high mutation frequency has never been reported for B. subtilis.
Role of chromosomal location and direct repeat for the high reversion frequency of gudB.
The extremely high frequency of reversion of gudB might result from the presence of a direct repeat of 9 bp. However, the chromosomal arrangement might play a role as well. To distinguish between these possibilities, we decided to address the role of the chromosomal location of the gudB gene first. For this purpose, we used the B. subtilis strain GP1163. In this strain, the chromosomal gudB gene was deleted, and another copy of gudB under the control of its own promoter was inserted ectopically at the amyE site of the chromosome. The ectopic copy of the gudB allele was orientated in the same orientation as the native copy. The mutation frequency of this strain was 0.49 × 10−4. Sequence analysis of three randomly selected suppressor mutants revealed that all contained the gudB1 mutation. This observation suggests that the chromosomal location has no major impact on the occurrence of the gudB1 suppressor mutation (see Table 3).
Table 3.
Frequency of gudB1 mutation
| Strain | Relevant genotype | Mutation frequency (avg ± SD)a |
|---|---|---|
| GP747 | rocG::Tn10 | (1.3 × 10−4) ± (40 × 10−4) |
| GP754 | rocG::cat | (1.1 × 10−4) ± (0.9 × 10−4) |
| GP1103 | rocG::Tn10 recA | (0.3 × 10−4) ± (0.4 × 10−4) |
| GP1107 | rocG::cat ΔaddAB | (0.9 × 10−4) ± (0.4 × 10−4) |
| GP1123 | amyE::(alf-aphA3 lacZ cat) | (1.3 × 10−7) ± (0.61 × 10−7) |
| GP1163 | rocG::Tn10 ΔgudB::aphA3 amyE::(gudB cat) | (0.49 × 10−4) ± (0.4 × 10−4) |
| GP1168 | amyE::(alf-aphA3 lacZ cat) Δmfd::ermC | (2.3 × 10−7) ± (0.48 × 10−7) |
| GP1169 | rocG::Tn10 Δmfd::ermC | 1.0 × 10−6b |
| GP1176 | rocG::Tn10 ΔuvrAB::ermC | (0.1 × 10−4) ± (0.02 × 10−4) |
| GP1179 | rocG::Tn10 ΔgudB::aphA3 amyE::(gudBmut1 cat) | (3.6 × 10−6) ± (2.7 × 10−6) |
| GP1180 | rocG::Tn10 ΔgudB::aphA3 amyE::(gudBmut2 cat) | (0.89 × 10−4) ± (0.14 × 10−4) |
| GP1191 | rocG::Tn10 ΔmutSL::aphA3 | (0.2 × 10−4) ± (0.07 × 10−4) |
| GP1192 | rocG::Tn10 ΔmutSL::aphA3 ΔuvrAB::ermC | (0.14 × 10−4) ± (0.03 × 10−4) |
| GP1197 | rocG::Tn10 ΔgudB::aphA3 amyE::(gudBmut3 cat) | (3.4 × 10−6) ± (8 × 10−6) |
Mutation frequencies were determined at least three times.
Due to the low mutation frequency of the gudB allele in GP1169, the determination of the precise frequency was limited by the experimental procedure.
As shown above, the gudB gene can be decryptified by the deletion of one part of the direct repeat irrespective of the chromosomal location of the gudB allele. Next, we wanted to address the relevance of this repeat in the decryptification process by a mutational analysis. Since the direct repeat is located within the coding sequence of gudB, any mutation to be introduced into the direct repeat had to conserve the gudB open reading frame. In order to destroy the direct repeat, we replaced two G residues by T (at positions 3 and 9 of the repeat, corresponding to wobble bases of the codons for valine and alanine). This mutation was introduced into both the first part and the second part of the direct repeat; the corresponding strains are B. subtilis GP1179 and GP1197, respectively. Moreover, we restored a direct repeat, albeit with a sequence that deviates from the original repeat by introducing the same mutations in both parts of the repeat. This strain was GP1180 (Fig. 1A). A comparison of the mutation frequencies revealed that the perfect repeat was a prerequisite for efficient accumulation of gudB1 suppressor mutants. In the absence of a perfect direct repeat, the mutation frequency was reduced by a factor of about 15 (0.036 × 10−4 and 0.034 × 10−4 for GP1179 and GP1197, respectively, versus 0.49 × 0−4 for strain GP1163 carrying the wild-type repeat; see Table 3). The introduction of compensatory mutations that restore the direct repeat did also restore the high frequency of the appearance of the gudB1 mutation (0.89 × 10−4 for GP1180). These results clearly demonstrate that the presence of the direct repeat is the decisive factor for the high gudB1 mutation frequency.
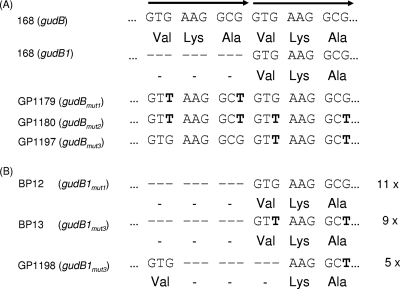
Fig 1.
Crucial role of the direct repeat for the decryptification of gudB. (A) The wild-type gudB sequence was mutated without changing the amino acid sequence. In GP1179 two G residues were replaced by T in the first half of the repeat (positions 3 and 9 of the repeat). In GP1197 these mutations were introduced in the second part of the direct repeat (positions 12 and 18). The perfect direct repeat was restored in the strain GP1180. This study served to analyze the role of a perfect direct repeat in the rapid decryptification of the gudB allele. (B) Selective deletion of the first part of the direct repeat in the gudB gene. In all gudB1 mutants (designated as BP12) derived from the strain GP1179, the first part of the imperfect repeat was excised. Of 14 gudB1 mutants derived from the strain GP1197, in 9 (designated as BP13) the first half of the imperfect repeat was deleted, whereas in 5 (designated as GP1198) an internal excision had occurred. The numbers indicate the number of occurrences of the particular mutations in a selected set of mutants that were analyzed.
Selective excision of the first part of the direct repeat.
In all experiments to determine mutation frequencies, we analyzed the nucleotide sequence of the gudB suppressor mutations. As stated above, a precise deletion of the direct repeat was observed in all cases. However, the presence of a perfect repeat precluded the identification of the nucleotides that had actually been excised. This question became tractable with the availability of the suppressor mutants of B. subtilis GP1179 and GP1197 in which the repeat is not perfect. The sequence analysis of the gudB1 alleles of 11 suppressor mutants (designated as BP12) derived from GP1179 (mutated in the first part of the repeat) revealed that the first half of the repeat was deleted in all cases (see Fig. 1B). This strong bias might indicate that either the first part of the repeat is preferentially excised or that the naturally occurring sequence is retained with preference. This question was addressed by analysis of the suppressor mutants derived from GP1197 (mutations in the second part of the repeat). In this case, of 14 analyzed mutants, 9 (designated as BP13) had a deletion of the first part of the repeat. Moreover, five mutants (designated as GP1198) exhibited internal deletions of the repeat that restored a sequence coding for the active GudB protein (see Fig. 1B). Thus, none of the mutants derived from GP1197 restored the original nucleotide sequence of the remainder of the repeat. Instead, we observed again a strong bias toward deletion of the first part of the repeat, suggesting that this selective deletion is inherent to the mutagenesis process that decryptifies the gudB gene.
Construction and analysis of a deletion reporter system.
The results presented above demonstrate that the deletion of one part of the gudB repeat occurs at a very high frequency both in the native context and in a nonrelated genomic context as long as the repeat is intact. These findings prompted us to ask whether the deletion would also take place as efficiently in a completely different sequence context as it does in the gudB gene. For this purpose, we constructed a reporter system consisting of an aphA3-lacZ operon encoding a resistance to kanamycin and β-galactosidase under the control of an artificial (alf) promoter. This promoter was designed to have perfect recognition sequences for the housekeeping sigma factor of the RNA polymerase (−10 and −35); however, the spacing between the two boxes was 26 bp rather than the canonical 17 to 18 bp. The perfect repeat of the gudB gene should be a part of this spacer. This promoter is not likely to be recognized by the RNA polymerase unless one part of the repeat is deleted and the optimal 17-bp spacing is restored (Fig. 2). Such a reporter system was constructed as described in Materials and Methods and introduced into the genome of B. subtilis, resulting in strain GP1123 (Table 1). B. subtilis GP1123 was unable to grow in the presence of kanamycin and formed white colonies on plates containing X-Gal, suggesting that neither kanamycin resistance nor β-galactosidase was expressed by these bacteria. These findings demonstrate that the alf promoter was inactive, as expected. However, we observed the sporadic appearance of kanamycin-resistant blue colonies that might result from the activation of the alf promoter. Indeed, a sequence analysis of the promoter for several colonies revealed the deletion of one part of the repeat resulting in a promoter (alf1) with perfect −10 and −35 regions separated by the preferred distance of 17 bp. Thus, the deletion of one part of the repeat occurs also in an unrelated sequence context.
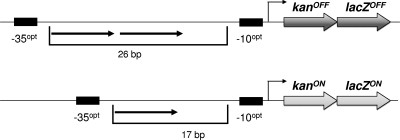
Fig 2.
Mutagenesis test system. The direct repeat originating from the gudB allele of B. subtilis was placed as the spacer between an optimal −10 and −35 region (upper part). An operon consisting of a kanamycin resistance gene (aphA3) and the β-galactosidase gene (lacZ) was placed under the control of the artificial promoter. Due to the long spacer, the promoter is not active. By the precise deletion of 9 bp in the spacer region, the promoter gains function, and the kanamycin resistance and the β-galactosidase are highly expressed (lower part).
Next, we sought to determine whether the deletion of the repeat in the alf promoter took place with a similar high frequency as observed for the decryptification of gudB. For this purpose, the frequency of appearance of kanamycin-resistant suppressor mutants of GP1123 was determined. It was found to be 1.3 × 10−7. This mutation frequency is in the range typically observed in bacteria (31), but 3 orders of magnitude lower than the frequency found for the deletion event in the gudB gene context. Thus, there seems to be a relevant difference between the sequence contexts of the gudB gene and the alf promoter that results in drastically changed mutation frequencies (see Table S2 in the supplemental material).
Expression of gudB gene and stability of cryptic and active glutamate dehydrogenases.
While the direct repeat is part of a putatively expressed coding region in the gudB gene, it is present in the nontranscribed spacer in the artificial alf promoter. This difference might contribute to the different mutation frequencies observed in the two sequence contexts. Therefore, we decided to study first the expression of the gudB gene to some detail. Previous studies have shown that gudB expression is not modulated by the source of nitrogen present in the medium (5). We have studied the activity of the gudB promoter by determining the expression of a gudB-lacZ fusion in wild-type, ΔrocG, and gudB1 genetic backgrounds. As shown in Table 2, the fusion was highly expressed irrespective of the genetic background or medium analyzed. The expression level of about 500 U/mg of protein is rather high for translational lacZ reporter fusions (50). Thus, even the cryptic gudB gene coding for an inactive protein is expressed at high levels in B. subtilis.
Table 2.
Analysis of gudB expression
| Strain | Relevant genotype | β-Galactosidase activity (U mg of protein−1)a |
||||
|---|---|---|---|---|---|---|
| C-Glc | CE | CE-Glc | CR | CR-Glc | ||
| GP1101 | Wild type | 357 | NG | 504 | 573 | 415 |
| GP1102 | gudB1 | 182 | 384 | 268 | 415 | 242 |
| GP1104 | rocG::Tn10 | 422 | NG | 557 | NG | 394 |
| GP1105 | rocG::Tn10 gudB1 | 225 | 658 | 478 | 410 | 295 |
Bacteria were grown in C minimal medium. Glucose (Glc), glutamate (E), and arginine (R) were added to final concentrations of 0.5% (Glc and R) or 0.8% (E). Experiments were carried out at least 3-fold. The maximum deviation of the series of representative data shown here was <30%. NG, no growth.
To allow the action of selective pressure on the decryptification of gudB, the accumulation of the active protein is required. However, the inactive GudB protein was reported to be one of the most unstable proteins of B. subtilis (23). In contrast, preliminary evidence suggested that the active GudB1 protein is much more stable (25). The issue of stability might apply not only at the level of the protein, but may also be relevant for the gudB mRNA. To address these problems, we first determined the stability of the gudB mRNA of B. subtilis GP747 and the isogenic gudB1 mutant GP753 by a Northern blot analysis. As shown in Fig. 3A, we detected a single transcript of ∼1,300 bp for gudB. This corresponds to a monocistronic transcript and is in good agreement with previous suggestions based on genome analysis (5). The quantitative evaluation of the mRNA stability revealed a half-life of ∼4 min. The stability of the mRNA was similar in both strains, demonstrating that it is not affected by the presence of the direct repeat.
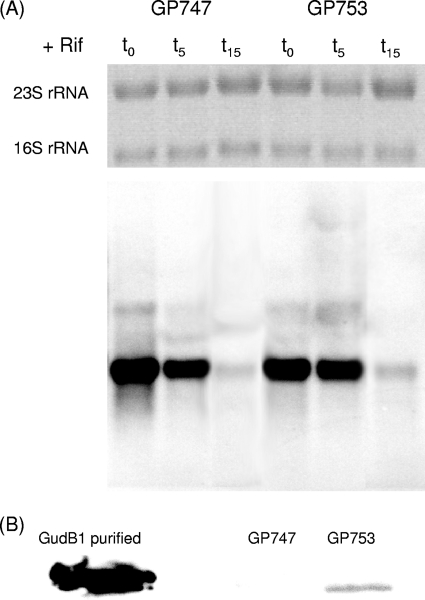
Fig 3.
Expression of gudB gene and GudB protein level. (A) Northern blot analysis was performed to determine the stability of the gudB mRNA of B. subtilis GP747 and the isogenic gudB1 mutant GP753. Both mRNAs do not differ in stability and half-life, implying that the direct repeat does not influence mRNA levels. (B) A Western blot analysis was performed to compare the protein levels of GudB with the level of GudB1. Crude extracts of B. subtilis GP747 and the isogenic gudB1 mutant GP753 were used, and the GudB protein was detected by using antibodies raised against RocG that cross-react with the GudB protein.
The accumulation of the glutamate dehydrogenase GudB was studied by Western blot analysis. For this purpose, we used the cell extracts of the rocG mutant GP747 and its isogenic gudB1 derivative GP753 that were prepared for the determination of the mRNA stability (just prior to rifampin addition, lane t0 in Fig. 3A). To detect the GudB protein, we used an antibody raised against RocG. Both proteins are very similar and the antibody recognizes GudB as well. Since both strains used for this experiment are rocG mutants, the only signal is obtained with GudB (17). As shown in Fig. 3B, the active enzyme GudB1 could be detected in the extract of GP753. In contrast, no signal was observed for the cryptic GudB protein. Since the mRNA amounts are similar for both strains (see Fig. 3A), we may conclude that the inactive GudB protein is highly unstable, as suggested by a previous study (23). In contrast, the active glutamate dehydrogenase GudB1 is a stable protein that accumulates in the cell. Thus, the decryptification of gudB is sufficient for the cell to obtain immediately an active glutamate dehydrogenase that may help to overcome the metabolic imbalance of the rocG mutant.
Implication of repair and recombination proteins in the decryptification of gudB.
The extremely high frequency at which the gudB decryptification occurs and the fact that the frequency is much higher in the gudB locus compared to the alf mutagenesis reporter system suggest the involvement of proteins in the mutagenesis process. The sequence of the direct repeat in gudB is somewhat similar to the chi sequence that is recognized and bound by the AddAB helicase/nuclease, a component of the recombination machinery of the cell. Since the recombination protein RecA is also involved in the generation of mutations, we determined the gudB mutation frequency of the addAB and recA mutant strains GP1107 and GP1103, respectively. The frequencies were similar to those observed with the isogenic rocG mutants (0.9 × 10−4 and 1.1 × 10−4 for the addAB mutant and the wild-type strain GP754; 0.3 × 10−4 versus 1.3 × 10−4 for the recA mutant and the wild-type strain GP747; Table 3). Therefore, AddAB and RecA do not seem to play a major role in the deletion of the direct repeat in the gudB gene. We also tested the effect of mutations in the genes recJ, recU, exoA, nfo, uvrAB, mutSL, and sbcDC. Moreover, we tested the effect of the combined exoA nfoA (GP1504) and mutSL uvrAB (GP1192) mutations. Similarly to the addAB and recA deletions, we did not observe an effect of the mutations on the decryptification of the gudB gene (see Table 3).
If a mutation in any of the genes encoding enzymes of DNA repair and recombination would have played a role in the deletion of the direct repeat in gudB, we would have expected that they are not selective for the gudB gene context compared to the context of the alf promoter. Thus, the genetic context plays a decisive role in the decryptification of gudB. As shown above, the gudB gene is constitutively expressed. In contrast, the core promoter of the mutagenesis reporter system is a nontranscribed region. The transcription-repair coupling factor Mfd might therefore participate in the deletion of the gudB repeat. To test this idea, we constructed the mfd deletion mutant GP1169 and compared the mutation frequency in this strain to that of the isogenic rocG mutant GP747. In this case, we detected a 100-fold reduction in the frequency of gudB1 mutants (1.25 × 10−6 versus 1.3 × 10−4). Next, we investigated the impact of the mfd mutation on the deletion of the repeat in the alf promoter. In this case, the mutation frequencies of the wild-type strain (GP1123) and the isogenic mfd mutant strain GP1168 were very similar (1.3 × 10−7 versus 2.3 × 10−7). Thus, the mfd mutation affects the deletion of the direct repeat only in the context of the transcribed gene. This observation strongly supports the idea that transcription of the gudB gene is essential for obtaining the high frequency of decryptification.
DISCUSSION
High-fidelity DNA synthesis is very important for maintaining genetic information over many generations of a bacterial population. Indeed, the frequency of single base pair substitutions during DNA replication is very low in E. coli. The frequency of these mutagenic events was estimated to be in the range of 10−7 to 10−8 in the absence of internal or external stress (49). Beneficial mutations occur even 2 orders of magnitudes less frequently (27, 29, 31). However, research in the last few years suggests that selective pressure may somehow favor the appearance of beneficial mutations (13, 47). A recent long-term study with E. coli suggested that the cells acquire the most beneficial mutations early during starvation (i.e., mutations that have the highest positive impact on fitness) and that independent bacterial cultures are likely to accumulate the same beneficial mutations (3). However, in contrast to base pair substitutions that occur during DNA synthesis, the occurrence of other genetic events such as transpositions, RecA-dependent deletions and inversions may vary from moderately frequent to very frequent.
The gudB1 mutation studied here appeared with a frequency of about 10−4. To the best of our knowledge, this is the highest mutation frequency for a specific allele that has been observed in B. subtilis. A particular feature of the gudB gene is the presence of a tandem repeat. As shown in the present study, the tandem repeat is essential for the high frequency of gudB decryptification, and any mutation that impairs the integrity of one of the tandem repeat units resulted in a reduced mutation frequency.
In bacteria, tandem repeats, often termed contingency loci, can be located within an open reading frame or in promoter regions (41). The longest tract of 57 tandem repeats was identified in the licA1 gene of H. influenzae (26). Moreover, tandem repeats are usually very instable since they are prone to high frequencies of mutations through slipped DNA strand mispairing (11, 41, 53). Well-studied tandem repeats such as the lgtC repeat in Haemophilus influenzae or the nadA repeat in Neisseria meningitidis are hot spots to generate phenotypic variation, thereby allowing the bacteria to adapt to changing environmental conditions (4, 36). The frequencies of the phenotypic variations range from 10−2 to 10−5 (41). Thus, the frequency of gudB decryptification is in the range observed for other tandem repeats.
In contrast to the well-studied examples, the tandem repeat in the gudB gene in the domesticated strain 168 possesses only two repeat units composed of nine nucleotides each. Moreover, the gudB repeat is surprisingly quite stable in a nonrelated, nontranscribed genomic context under laboratory growth conditions. Indeed, a derivative of the strain 168 with an active GudB glutamate dehydrogenase could only be selected on minimal medium with glutamate as the single carbon source (5, 19). The situation is completely different when the rocG gene that encodes the final enzyme of the arginine degradation pathway is inactivated. These bacteria form only small translucent colonies on complex medium and rapidly acquire the gudB1 mutation. The occurrence of the gudB1 mutation at such a high frequency suggests the existence of a strong selective pressure exerted on the rocG mutant. The rocG gene product, the glutamate dehydrogenase, converts glutamate to 2-oxoglutarate. This suggests that glutamate or one of its precursors in the arginine degradation pathway might accumulate in the rocG mutant, and this might be problematic for the cell. We have tested the growth of mutants affected in the different steps of arginine degradation on complex medium; however, the strong growth defect was unique to the rocG mutant (our unpublished results). Thus, the accumulation of glutamate may be toxic for the cell. This idea is in good agreement with the observation that a strain with a constitutive high-level expression of the glutamate synthesizing enzyme glutamate synthase acquired a mutation that inactivates this enzyme when grown in the presence of glutamate (19). This leaves us with the question why glutamate should be toxic for the cell when it is the most abundant metabolite anyway. The enzyme glutamate racemase (encoded by the essential gene racE in B. subtilis) catalyzes the conversion of l-glutamate to d-glutamate that is a building block for peptidoglycan biosynthesis (30, 52). Indeed, the accumulation d-glutamate was shown to be toxic for B. subtilis (30). In the presence of very high intracellular amounts of l-glutamate due to the strong induction of the enzymes of the arginine degradation pathway, RacE probably generates higher concentrations of d-glutamate than are tolerated by the cell. The activation of the normally cryptic glutamate dehydrogenase GudB might then bring the glutamate concentration to a level that does not longer result in the accumulation of harmful d-glutamate.
The high frequency and the high precision of the gudB decryptification imply that the molecular tools to generate the mutation must be present in B. subtilis. It has been reported that tandem repeat deletions occur in E. coli rather during chromosome replication by slipped DNA strand mispairing than via the RecA-dependent homologous recombination pathway (10, 11). This observation is in good agreement with our results that the mutations in the addAB and recA genes do not influence the decryptification of the gudB gene. Similarly, the absence of the proteins RecJ, RecU, ExoA, Nfo, UvrAB, MutSL, and SbcDC that are involved in DNA repair, recombination, and stationary-phase mutagenesis did not affect the high-frequency gudB mutation. However, we cannot exclude that other factors involved in the excision of the tandem repeat in the gudB gene escaped our attention. Moreover, the observed bias for selective excision of the first part of the direct repeat might be explained by the slipped DNA strand mispairing model (11). To the best of our knowledge, this was not yet shown for tandem repeat deletions in bacteria. Thus far, there are no reports available that describe the underlying molecular mechanism and the players involved in tandem repeat deletions in B. subtilis and any other Gram-positive species. However, short sequence tandem repeats may play a role in the adaptation of clinical isolates of Streptococcus pneumoniae as a result of selective pressure exerted by the human immune system (43).
Mfd is a multifunctional protein that can play different roles in the cell, e.g., Mfd is involved in transcription-coupled DNA repair and the removal of stalled transcription complexes from DNA (12). However, previous observations indicate that Mfd may facilitate the acquisition of beneficial mutations in the stationary growth phase in B. subtilis (45, 46). Our work shows that the Mfd protein is essential for the high-frequency decryptification of the gudB gene. The decryptification of the gene occurs by deleting one part of a direct repeat that is located in a transcribed region. The identification of other enzymes that are required for the decryptification of gudB and the underlying molecular mechanism will be the subject of further analyses.
The Mfd-mediated coupling of transcription to DNA repair and mutagenesis can be regarded as a built-in precaution that facilitates the accumulation of mutations preferentially in transcribed genes. This has several implications. (i) The coupling allows that the mutations occur in genes that are expressed at the given time point; therefore, the mutant variants of the encoded proteins might help to overcome the actual limitation. (ii) Nontranscribed genes that may be required under different conditions are in this way protected from potentially harmful mutations. Both effects facilitate the adaptation of bacteria to all kind of challenges that limit their growth and are therefore crucial for bacterial evolution.
ACKNOWLEDGMENTS
Rafik Neme Garrido, Kristin Kaiser, and Bernard Freytag are acknowledged for their help with some experiments. We thank Frederik M. Meyer, Marie-Francoise Noirot-Gros, Hinnerk Eilers, and Wilfried Kramer for helpful discussions. We are grateful to Chiara Marchisone, Dusko Ehrlich, and Alan Grossman for providing strains BG427, HVS666, and IRN444, respectively.
This study was supported by the DFG, the Federal Ministry of Education (Research SYSMO network [PtJ-BIO/0313978D]), and the Fonds der Chemischen Industrie (J.S.).
Footnotes
Published ahead of print 16 December 2011
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Abramoff MD, Magelhaes PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics Internat. 11:36–42 [Google Scholar]
- 2. Ayora S, Rojo F, Ogasawara N, Nakai S, Alonso JC. 1996. The Mfd protein of Bacillus subtilis 168 is involved in both transcription-coupled DNA repair and DNA recombination. J. Mol. Biol. 256:301–318 [DOI] [PubMed] [Google Scholar]
- 3. Barrick JE, et al. 2009. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461:1243–1247 [DOI] [PubMed] [Google Scholar]
- 4. Bayliss CD, Field D, Moxon ER. 2001. The simple sequence contingency loci of Haemophilus influenzae and Neisseria meningitidis. J. Clin. Invest. 107:657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Belitsky BR, Sonenshein AL. 1998. Role and regulation of Bacillus subtilis glutamate dehydrogenase genes. J. Bacteriol. 180:6298–6305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Belitsky BR, Sonenshein AL. 1999. An enhancer element located downstream of the major glutamate dehydrogenase gene of Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 96:10290–10295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Belitsky BR, Kim HJ, Sonenshein AL. 2004. CcpA-dependent regulation of Bacillus subtilis glutamate dehydrogenase gene expression. J. Bacteriol. 186:3392–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Belitsky BR, Sonenshein AL. 2004. Modulation of activity of Bacillus subtilis regulatory proteins GltC and TnrA by glutamate dehydrogenase. J. Bacteriol. 186:3399–3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bi W, Stambrook PJ. 1998. Site-directed mutagenesis by combined chain reaction. Anal. Biochem. 256:137–140 [DOI] [PubMed] [Google Scholar]
- 10. Bichara M, Wagner J, Lambert IB. 2006. Mechanisms of tandem repeat instability in bacteria. Mutat. Res. 598:144–163 [DOI] [PubMed] [Google Scholar]
- 11. Bierne H, Vilette D, Ehrlich SD, Michel B. 1997. Isolation of dnaE mutation which enhances RecA-independent homologous recombination in the Escherichia coli chromosome. Mol. Microbiol. 24:1225–1235 [DOI] [PubMed] [Google Scholar]
- 12. Borukhov S, Lee J, Laptenko O. 2005. Bacterial transcription elongation factors: new insights into molecular mechanism of action. Mol. Microbiol. 55:1315–1342 [DOI] [PubMed] [Google Scholar]
- 13. Cairns J, Overbaugh J, Miller S. 1988. The origin of mutants. Nature 335:142–145 [DOI] [PubMed] [Google Scholar]
- 14. Chédin F, Noirot P, Biaudet V, Ehrlich SD. 1998. A five-nucleotide sequence protects DNA from exonucleolytic degradation by AddAB, the RecBCD analogue of Bacillus subtilis. Mol. Microbiol. 29:1369–1377 [DOI] [PubMed] [Google Scholar]
- 15. Commichau FM, Forchhammer K, Stülke J. 2006. Regulatory links between carbon and nitrogen metabolism. Curr. Opin. Microbiol. 9:167–172 [DOI] [PubMed] [Google Scholar]
- 16. Commichau FM, Herzberg C, Tripal P, Valerius O, Stülke J. 2007. A regulatory protein-protein interaction governs glutamate biosynthesis in Bacillus subtilis: the glutamate dehydrogenase RocG moonlights in controlling the transcription factor GltC. Mol. Microbiol. 65:642–654 [DOI] [PubMed] [Google Scholar]
- 17. Commichau FM, et al. 2007. Characterization of Bacillus subtilis mutants with carbon source-independent glutamate biosynthesis. J. Mol. Microbiol. Biotechnol. 12:106–113 [DOI] [PubMed] [Google Scholar]
- 18. Commichau FM, Stülke J. 2008. Trigger enzymes: bifunctional proteins active in metabolism and in controlling gene expression. Mol. Microbiol. 67:692–702 [DOI] [PubMed] [Google Scholar]
- 19. Commichau FM, Gunka K, Landmann JJ, Stülke J. 2008. Glutamate metabolism in Bacillus subtilis: gene expression and enzyme activities evolved to avoid futile cycles and to allow rapid responses to perturbations of the system. J. Bacteriol. 190:3557–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fernández S, Sorokin A, Alonso JC. 1998. Genetic recombination in Bacillus subtilis 168: effects of recU and recS mutations on DNA repair and homologous recombination. J. Bacteriol. 180:3405–3409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Flórez LA, Gunka K, Polanía R, Tholen S, Stülke J. 2011. SPABBATS: a pathway-discovery method based on Boolean satisfiability facilitates the characterization of suppressor mutants. BMC Syst. Biol. 5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fukui K. 2010. DNA mismatch repair in eukaryotes and bacteria. J. Nucleic. Acids. pii: 260512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gerth U, et al. 2008. Clp-dependent proteolysis downregulates central metabolic pathways in glucose-starved Bacillus subtilis. J. Bacteriol. 190:321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guérout-Fleury AM, Shazand K, Frandsen N, Stragier P. 1995. Antibiotic resistance cassettes for Bacillus subtilis. Gene 167:335–336 [DOI] [PubMed] [Google Scholar]
- 25. Gunka K, et al. 2010. Functional dissection of a trigger enzyme: mutations of the Bacillus subtilis glutamate dehydrogenase RocG that affect differentially its catalytic activity and regulatory properties. J. Mol. Biol. 400:815–827 [DOI] [PubMed] [Google Scholar]
- 26. High NJ, Jennings MP, Moxon ER. 1996. Tandem repeats of the tetramer 5′-CAAT-3′ present in licA2 are required for the phase variation but not lipopolysaccharide biosynthesis in Haemophilus influenzae. Mol. Microbiol. 20:165–174 [DOI] [PubMed] [Google Scholar]
- 27. Imhof M, Schlötterer C. 2001. Fitness effects of advantageous mutations in evolving Escherichia coli populations. Proc. Natl. Acad. Sci. U. S. A. 98:1113–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kempf B, Bremer E. 1998. Uptake and synthesis of compatible solutes as microbial stress responses to high osmolarity environments. Arch. Microbiol. 170:319–330 [DOI] [PubMed] [Google Scholar]
- 29. Kibota TT, Lynch M. 1996. Estimate of the genomic mutation frequency deleterious to overall fitness in Escherichia coli. Nature 381:694–696 [DOI] [PubMed] [Google Scholar]
- 30. Kimura K, Tran L-SP, Itoh Y. 2004. Roles and regulation of the glutamate racemase isogenes, racE and yrpC, in Bacillus subtilis. Microbiology 150:2911–2920 [DOI] [PubMed] [Google Scholar]
- 31. Kunkel TA. 2004. DNA replication fidelity. J. Biol. Chem. 279:16895–16898 [DOI] [PubMed] [Google Scholar]
- 32. Kunst F, Rapoport G. 1995. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol. 177:2403–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lea DE, Coulson CA. 1949. The distribution of mutants in bacterial populations. J. Genet. 49:248–264 [DOI] [PubMed] [Google Scholar]
- 34. Lemon KP, Kurtser I, Grossman AD. 2001. Effects of replication termination mutants on chromosome partitioning in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 98:212–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ludwig H, et al. 2001. Transcription of glycolytic genes and operons in Bacillus subtilis: evidence for the presence of multiple levels of control of the gapA operon. Mol. Microbiol. 41:409–422 [DOI] [PubMed] [Google Scholar]
- 36. Martin P, Makepeace K, Hill SA, Hood DW, Moxon ER. 2005. Microsatellite instability regulates transcription factor binding and gene expression. Proc. Natl. Acad. Sci. U. S. A. 102:3800–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Martin-Verstraete I, Débarbouillé M, Klier A, Rapoport G. 1992. Mutagenesis of the Bacillus subtilis “-12,-24” promoter of the levanase operon and evidence for the existence of an upstream activating sequence. J. Mol. Biol. 226:85–99 [DOI] [PubMed] [Google Scholar]
- 38. Meinken C, Blencke HM, Ludwig H, Stülke J. 2003. Expression of the glycolytic gapA operon in Bacillus subtilis: differential syntheses of proteins encoded by the operon. Microbiology 149:751–761 [DOI] [PubMed] [Google Scholar]
- 39. Meyer FM, et al. 2011. Physical interactions between tricarboxylic acid cycle enzymes in Bacillus subtilis: evidence for a metabolon. Metab. Eng. 13:18–27 [DOI] [PubMed] [Google Scholar]
- 40. Modrich P, Lahue R. 1996. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 65:101–133 [DOI] [PubMed] [Google Scholar]
- 41. Moxon R, Bayliss C, Hood D. 2006. Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Annu. Rev. Genet. 40:307–333 [DOI] [PubMed] [Google Scholar]
- 42. Oh YK, Palsson BO, Park SM, Schilling CH, Mahadevan R. 2007. Genome-scale reconstruction of metabolic network in Bacillus subtilis based on high-throughput phenotyping and gene essentiality data. J. Biol. Chem. 282:28791–28799 [DOI] [PubMed] [Google Scholar]
- 43. Pericone CD, Bae D, Shchepetov M, McCool T, Weiser JN. 2002. Short-sequence tandem and nontandem DNA repeats and endogenous hydrogen peroxide production contribute to genetic instability of Streptococcus pneumoniae. J. Bacteriol. 184:4392–4399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Picossi S, Belitsky BR, Sonenshein AL. 2007. Molecular mechanism of the regulation of Bacillus subtilis gltAB expression by GltC. J. Mol. Biol. 365:1298–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pybus C, et al. 2010. Transcription-associated mutation in Bacillus subtilis cells under stress. J. Bacteriol. 192:3321–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ross C, et al. 2006. Novel role of mfd: effects on stationary-phase mutagenesis in Bacillus subtilis. J. Bacteriol. 188:7512–7520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Roth JR, Kugelberg E, Reams AB, Kofoid E, Andersson DI. 2006. Origin of mutations under selection: the adaptive mutation controversy. Annu. Rev. Genet. 60:477–501 [DOI] [PubMed] [Google Scholar]
- 48. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 49. Schaaper RM. 1993. Base selection, proofreading, and mismatch repair during DNA replication in Escherichia coli. J. Biol. Chem. 286:23762–23765 [PubMed] [Google Scholar]
- 50. Schilling O, et al. 2007. Transcriptional and metabolic responses of Bacillus subtilis to the availability of organic acids: transcription regulation is important but not sufficient to account for metabolic adaptation. Appl. Environ. Microbiol. 73:499–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sonenshein AL. 2007. Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 5:917–927 [DOI] [PubMed] [Google Scholar]
- 52. Spies MA, et al. 2009. Determinants of catalytic power and ligand binding in glutamate racemase. J. Am. Chem. Soc. 131:5274–5284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Streisinger G, et al. 1966. Frameshift mutations and the genetic code. Cold Spring Harbor Symp. Quant. Biol. 31:77–84 [DOI] [PubMed] [Google Scholar]
- 54. Stülke J, et al. 1997. Induction of the Bacillus subtilis ptsGHI operon by glucose is controlled by a novel antiterminator, GlcT. Mol. Microbiol. 25:65–78 [DOI] [PubMed] [Google Scholar]
- 55. Sung HM, Yasbin RE. 2002. Adaptive, or stationary-phase, mutagenesis, a component of bacterial differentiation in Bacillus subtilis. J. Bacteriol. 184:5641–5653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Truglio JJ, Croteau DL, Van Houten B, Kisker C. 2006. Prokaryotic nucleotide excision repair: the UvrABC system. Chem. Rev. 106:233–252 [DOI] [PubMed] [Google Scholar]
- 57. Veening JW, Kuipers OP. 2010. Gene position within a long transcript as a determinant for stochastic switching in bacteria. Mol. Microbiol. 76:269–272 [DOI] [PubMed] [Google Scholar]
- 58. Wacker I, et al. 2003. The regulatory link between carbon and nitrogen metabolism in Bacillus subtilis: regulation of the gltAB operon by the catabolite control protein CcpA. Microbiology 149:3001–3009 [DOI] [PubMed] [Google Scholar]
- 59. Yan D. 2007. Protection of the glutamate pool concentration in enteric bacteria. Proc. Natl. Acad. Sci. U. S. A. 104:9475–9480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yuan J, et al. 2009. Metabolomics-driven quantitative analysis of ammonia assimilation in Escherichia coli. Mol. Syst. Biol. 5:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zeigler DR, et al. 2008. The origins of 168, W23, and other Bacillus subtilis legacy strains. J. Bacteriol. 190:6983–6995 [DOI] [PMC free article] [PubMed] [Google Scholar]