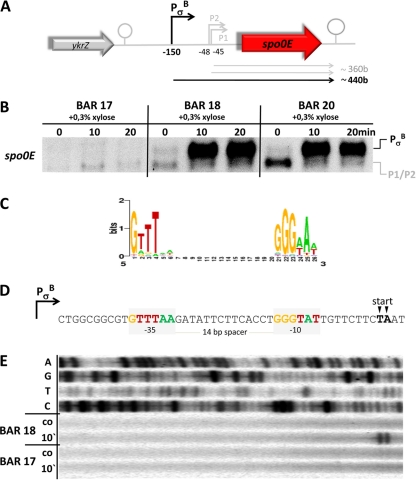
Fig 1.
Regulation of spo0E. (A) Schematic representation of the chromosomal spo0E region. (B) Northern blot experiment of the expression profiles of spo0E in response to addition of 0.3% (wt/vol) xylose to exponentially growing cultures of strains BAR 17 (ΔrsbVW-sigB), BAR 18 (ΔrsbVW-sigB amyE::PxylA-sigB), and BAR 20 (ΔrsbVW-sigB amyE::PxylA-sigB ΔabrB). Cells were grown in minimal media, and 0.3% xylose was added at t0 = OD500 of 0.4. Samples for RNA preparation were taken under control conditions before (0 min) as well as 10 and 20 min after addition of xylose. (C) Sequence logo of the σB consensus binding sites (http://dbtbs.hgc.jp/). The height of the letters in bits is proportional to their frequency (48). (D) Sequence of the σB promoter PσB of spo0E. The core promoter regions are marked with light gray boxes and bases matching the consensus were color coded accordingly. Transcriptional start points determined by the primer extension (PE) experiment are marked with arrows. (E) PE experiment to determine the 5′ end of the xylose-induced message observed in the Northern blot experiment. The upper 4 lines show dideoxy-sequencing ladders that were terminated with ddATP (line 1), ddGTP (line 2), ddTTP (line 3), and ddCTP (line 4). The lower 4 lines show reactions with RNA isolated from cells of the BAR 18 and BAR 17 strains under control conditions (co) before and 10 min (10′) after addition of 0.3% xylose.