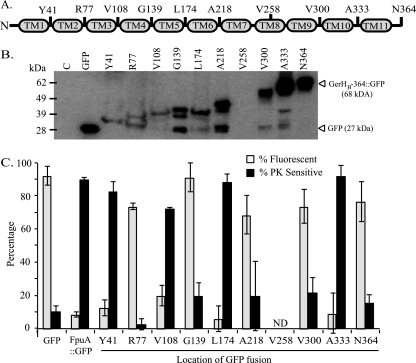
Fig 2.
Analysis of GerHB topology. (A) Model of GerHB with 11 putative transmembrane domains as determined by topology prediction. To test these putative TMs, C-terminal GFP fusions were made to full-length GerHB (N364) and nine GerHB proteins truncated at the indicated locations. TM8 was excluded because of its low predictive value. (B) Expression of GerHB fusion proteins in B. anthracis. Whole-cell lysates from bacteria expressing a vector control (lane C), GFP, or GerHB fusion proteins were analyzed by immunoblotting with anti-GFP antibodies to assess protein expression and stability. The predicted sizes of GFP and full-length GerHB-364::GFP are indicated. (C) Analysis of GerHB fusion proteins by flow cytometry and protease sensitivity assays. Flow cytometry was used to measure the fluorescence intensities of bacteria expressing control proteins (GFP or FpuA::GFP) or the GerHB fusion proteins. The percentage of GFP+ cells in each sample was determined as described for Fig. 1B (gray bars). Protease sensitivity assays were performed with proteinase K. The percentage of each fusion protein that was protease sensitive was determined as described for Fig. 1C (black bars). Each bar represents the mean ± standard error of the mean. ND, not determined.