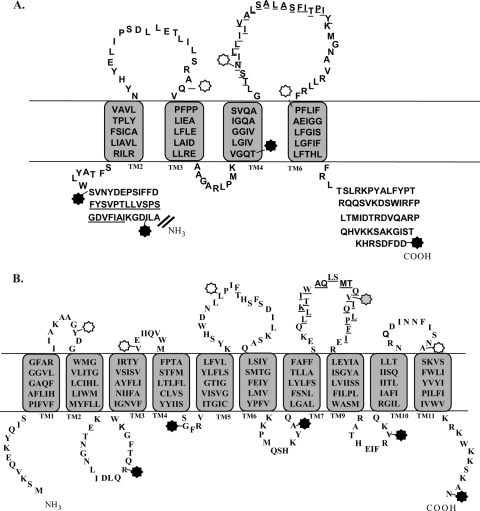
Fig 4.
Topology models of B. anthracis GerHA and GerHB. Topology models of the membrane-spanning regions (amino acids 490 to 747) of GerHA (A) and full-length GerHB (B) are based on a combination of experimental results and topology modeling. The locations of GFP fusions used in this study are marked by stars. Black stars represent fusion locations that were determined to be intracellular, while white stars represent extracellular fusion locations. The gray star marks the unstable GerHB fusion, which was unable to be analyzed. Membrane-spanning sequences were determined based on consensus sequences from 11 topology prediction algorithms. The actual membrane-spanning sequences of the proteins may vary from these predicted domains. Underlined amino acid residues indicate predicted transmembrane domains that were not supported by GFP fusion analysis.