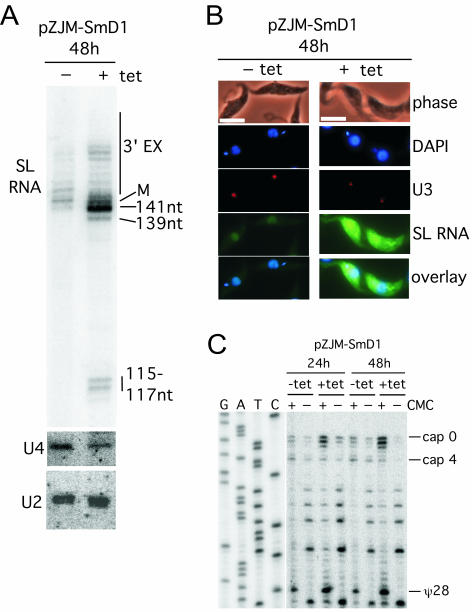
FIG. 2.
Knockdown of SmD1 mRNA affects spliced leader RNA abundance, size, and subcellular distribution. (A) High-resolution 6% acrylamide-8 M urea gels were used to separate RNA transcripts to the single-nucleotide level. Blots were probed with spliced leader (SL) RNA, U2, and U4 snRNA probes. M, mature size (142 nucleotides). (B) Fluorescence in situ hybridization analysis for intracellular localization of spliced leader RNA with a fluorescein isothiocyanate-labeled probe was performed on uninduced (without tetracycline, −tet) and induced (+tet) cells at 48 h. Bar, 10 μm. U3 snoRNA was localized with a Tamara-labeled oligonucleotide probe; nuclear and kinetoplast DNAs were stained with DAPI. (C) ψ28 occurs independent of SmD1. ψ28 was assayed on RNA from induced (+tet) and uninduced (−tet) populations with carboxymethyl cellulose (CMC) modification (+ or −) and primer extension. Products extended with the 5′-end-labeled TbSL stem-loop I oligonucleotide were resolved alongside a corresponding DNA sequencing ladder through a 6% acrylamide-8 M urea gel. Additional primer extension stops at three AU and one AC dinucleotide were seen inconsistently in different experiments.