Abstract
The genotypic diversity of Coxiella burnetii in clinical samples obtained from the Dutch Q fever outbreak episodes of 2007-2010 was determined by using a 6-locus variable-number tandem repeat analysis panel. The results are consistent with the introduction of one founder genotype that is gradually diversifying over time while spreading throughout The Netherlands.
TEXT
From 2007 to 2010, The Netherlands was confronted with a large and unprecedented Q fever outbreak, with thousands of affected individuals (4). The increase in human cases coincided with an increase in abortions among goats (2, 4, 6). Genotypic characterization of the involved isolates can give fundamental insight into the epidemiology of Q fever in The Netherlands, allowing, for example, spread of the involved genotype(s) throughout The Netherlands during the subsequent outbreak years and/or displaying a correlation between human and animal Q fever cases. Recently, genotyping by using a 10-locus multiple-locus variable-number tandem repeat analysis (MLVA) panel revealed one predominant genotype among goats and sheep throughout the affected area (5). A 3-locus MLVA panel performed directly on clinical samples from a minor part of the affected region showed that Dutch farm animals and patients appeared to be infected by different but closely related MLVA genotypes (3). In this study, we determined the temporal and spatial diversity of Coxiella burnetii genotypes in human samples collected during the 2007-2010 Q fever outbreak episodes from the entire affected part of The Netherlands using a 6-locus MLVA panel.
The presence of C. burnetii DNA in a variety of clinical samples was determined using a real-time PCR targeting the IS1111a insertion element of C. burnetii as described earlier (8). We determined the MLVA genotype using 3 hexanucleotide repeat markers (Ms27, Ms28, and Ms34) and 3 heptanucleotide repeat markers (Ms23, Ms24, and Ms33) (1) directly in 46 Q fever-positive clinical specimens collected from acute and chronic Q fever patients. These samples were collected during the 2007-2010 outbreak episodes (Table 1 and Fig. 1A). A multicolor multiplex format was chosen to make more efficient use of the small amounts of C. burnetii DNA generally obtained from clinical samples. The MLVA primers for markers Ms27, Ms28, and Ms34 have been described before (3). MLVA primers were 5′-HEX-CGCMTAGCGACACAACCAC-3′ and 5′-GACGGGCTAAATTACACCTGCT-3′ for Ms23, 5′-FAM-TGGAGGGACTCCGATTAAAA-3′ and 5′-GCCACACAACTCTGTTTTCAG-3′ for Ms24, and 5′-TAMRA-TCGCGTAGCGACACAACC-3′ and 5′-GTAGCCCGTATGACGCGAAC-3′ for Ms33, where HEX is hexachlorofluorescein, FAM is 6-carboxyfluorescein, and TAMRA is 6-carboxytetramethylrhodamine.
Table 1.
Clinical samples, geographical location, year of collection, and obtained MLVA genotypes from the Dutch Q fever outbreak episodes of 2007 to 2010a
| Sample no., strain, or source | Clinical specimen | Geographical location | Yr | CT value | No. of repeats |
MLVA type | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ms23 | Ms24 | Ms27 | Ms28 | Ms33 | Ms34 | ||||||
| Q012 | Urine | Balgoij | 2008 | 31.7 | 6 | 11 | 3 | 3 | 2 | 7 | A |
| Q013 | Sputum | Beneden-Leeuwen | 2008 | 31.9 | 6 | 11 | 3 | 3 | 2 | 7 | A |
| Q014 | Throat swab | Balgoij | 2008 | 31.9 | 6 | 11 | 3 | 3 | 2 | 7 | A |
| Q015 | Plasma | Grave | 2008 | 34.7 | 6 | 11 | 3 | 3 | 2 | 7 | A |
| Q032 | Serum | Wijchen | 2008 | 31.7 | 6 | 11 | 3 | 3 | 2 | 7 | A |
| Q033 | Serum | Nijmegen | 2007 | 30.9 | 6 | 11 | 3 | 3 | 2 | 7 | A |
| Q034 | BAL fluid | Wijk bij Duurstede | 2008 | 26.6 | 6 | 11 | 3 | 3 | 2 | 7 | A |
| Q007 | Throat swab | Alverna | 2008 | 31.9 | 6 | 11 | 3 | 3 | 2 | 8 | B |
| Q018 | Sputum | Balgoij | 2008 | 34.2 | 6 | 11 | 4 | 3 | 2 | 7 | C |
| Q008 | Plasma | Nijmegen | 2008 | 34.4 | 6 | 13 | 3 | 3 | 2 | 8 | D |
| Q042 | BAL fluid | Veldhoven | 2009 | 29.9 | 3 | 11 | 3 | 3 | 2 | 8 | E |
| Q084 | Aorta valve | Zeeland | 2008 | 17.0 | 3 | 11 | 3 | 3 | 2 | 8 | E |
| Q102 | Plasma | Nuenen | 2010 | 29.1 | 3 | 11 | 3 | 3 | 2 | 8 | E |
| Q072 | BAL fluid | Prinsenbeek | 2009 | 32.8 | 3 | 10 | 3 | 3 | 2 | 7 | F |
| Q050 | BAL fluid | Tilburg | 2009 | 22.4 | 3 | 11 | 3 | 3 | 2 | 7 | G |
| Q052 | Sputum | Houten | 2009 | 20.7 | 3 | 11 | 3 | 3 | 2 | 7 | G |
| Q053 | Sputum | Nieuwegein | 2009 | 23.3 | 3 | 11 | 3 | 3 | 2 | 7 | G |
| Q054 | Sputum | Houten | 2009 | 19.4 | 3 | 11 | 3 | 3 | 2 | 7 | G |
| Q055 | Sputum | Utrecht | 2009 | 26.0 | 3 | 11 | 3 | 3 | 2 | 7 | G |
| Q057 | Sputum | Houten | 2009 | 20.6 | 3 | 11 | 3 | 3 | 2 | 7 | G |
| Q063 | Sputum | Ravenstein | 2009 | 29.6 | 3 | 11 | 3 | 3 | 2 | 7 | G |
| Q064 | BAL fluid | Eindhoven | 2009 | 31.1 | 3 | 11 | 3 | 3 | 2 | 7 | G |
| Q066 | Sputum | Wijchen | 2009 | 27.7 | 3 | 11 | 3 | 3 | 2 | 7 | G |
| Q074 | Wound fluid | Groesbeek | 2009 | NA | 3 | 11 | 3 | 3 | 2 | 7 | G |
| Q076 | Aorta valve | Druten | 2009 | NA | 3 | 11 | 3 | 3 | 2 | 7 | G |
| Q078 | Aorta valve | Standaardbuiten | 2009 | 26.4 | 3 | 11 | 3 | 3 | 2 | 7 | G |
| Q083 | Thrombus | 's-Hertogenbosch | 2010 | 18.2 | 3 | 11 | 3 | 3 | 2 | 7 | G |
| Q099 | Abscess fluid | Son | 2010 | 24.6 | 3 | 11 | 3 | 3 | 2 | 7 | G |
| Q101 | Vascular prosthesis | Handel | 2010 | 22.7 | 3 | 11 | 3 | 3 | 2 | 7 | G |
| Q103 | Aorta tissue | Venlo | 2010 | 32.3 | 3 | 11 | 3 | 3 | 2 | 7 | G |
| Q104 | Serum | Wijchen | 2010 | 27.2 | 3 | 11 | 3 | 3 | 2 | 7 | G |
| Q107 | Aorta valve | Wijchen | 2010 | 9.0 | 3 | 11 | 3 | 3 | 2 | 7 | G |
| Goats (n = 20) | Placenta | Balgoij | 2008 | NA | 3 | 11 | 3 | 3 | 2 | 7 | G |
| Goat | Placenta | Wouda | 2009 | NA | 3 | 11 | 3 | 3 | 2 | 7 | G |
| Goat | Placenta | Denekamp | 2009 | NA | 3 | 11 | 3 | 3 | 2 | 7 | G |
| Q056 | BAL fluid | Amersfoort | 2009 | 28.2 | 4 | 11 | 3 | 3 | 3 | 8 | H |
| Q011 | Throat swab | Balgoij | 2008 | 31.7 | 0 | 11 | 3 | 3 | 0 | 7 | P |
| Q019 | Urine | Balgoij | 2008 | 36.8 | 0 | 11 | 0 | 3 | 0 | 4 | p |
| Q020 | Throat swab | Herpen | 2008 | 38.3 | 0 | 0 | 0 | 3 | 0 | 0 | p |
| Q021 | Throat swab | Nijmegen | 2008 | 37.8 | 0 | 0 | 4 | 3 | 0 | 0 | p |
| Q022 | Throat swab | Nijmegen | 2008 | 37.9 | 0 | 0 | 4 | 3 | 0 | 0 | p |
| Q067 | Serum | Tilburg | 2009 | 34.5 | 0 | 11 | 0 | 0 | 0 | 0 | p |
| Q070 | Serum | Tilburg | 2009 | 32.0 | 0 | 0 | 0 | 0 | 0 | 7 | p |
| Q075 | Serum | Druten | 2009 | NA | 0 | 11 | 0 | 3 | 0 | 0 | p |
| Q079 | Serum | Standaardbuiten | 2009 | NA | 0 | 11 | 3 | 3 | 0 | 0 | p |
| Q080 | Aorta tissue | Standaardbuiten | 2009 | NA | 0 | 11 | 3 | 3 | 0 | 0 | p |
| Q089 | Serum | Overloon | 2010 | 34.8 | 0 | 0 | 3 | 0 | 0 | 7 | p |
| Q098 | Abscess fluid | Son | 2010 | 28.3 | 0 | 11 | 3 | 3 | 0 | 7 | p |
| Q100 | Wound fluid | Handel | 2010 | 30.5 | 0 | 0 | 3 | 3 | 0 | 7 | p |
| C. burnetii Dugway | ND | 5 | 4 | 4 | 3 | 3 | |||||
| C. burnetii RSA 331 | 4 | 7 | 3 | 3 | −1b | 3 | |||||
| C. burnetii RSA 493 | DNA | 9 | 27 | 4 | 6 | 4 | 5 | ||||
| C. burnetii CbuG Q212 | ND | 8 | 3 | 4 | 2 | 2 | |||||
| C. burnetii CbuK Q154 | ND | 9 | 4 | 5 | 2 | 2 | |||||
The number of repeats in each marker was determined by extrapolation using the sizes of the obtained fragments relative to those obtained using DNA from the Nine Mile strain (RSA 493). Furthermore, the genotypes of four additional C. burnetii strains, i.e., Dugway (GenBank accession number CP000733), RSA331 (CP000890), CbuG Q212 (CP001019), and CbuK Q154 (CP001020) were determined in silico using the published sequences. NA, results not available; 0, no results obtained; p, partial genotype; ND, number of repeats could not be determined due to apparent sequence assembly errors.
In silico analysis showed 5 fewer repeats than the Nine Mile strain (RSA 493), which by convention was assigned 4 repeats (1).
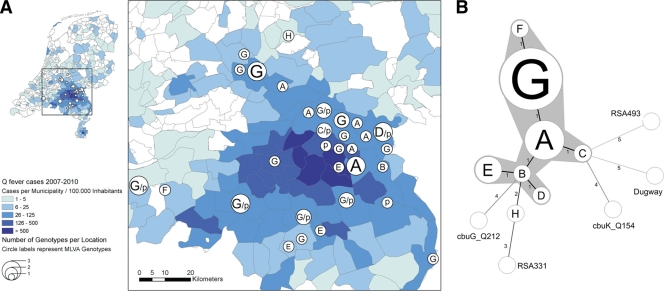
Fig 1.
Geographical distribution and relationship between Dutch C. burnetii genotypes from humans. (A) Geographical locations of the MLVA genotypes obtained from the 2007-2010 outbreak. Syllables correspond to the genotypes identified in Table 1. (B) Minimum spanning tree showing the relationship between the obtained MLVA genotypes identified in this study. Only full MLVA genotypes obtained from clinical samples were included in this analysis. Additionally, the genotypes from five sequenced C. burnetii strains, i.e., Dugway (GenBank accession number CP000733), RSA331 (CP000890), Nine Mile RSA493 (AE016828), CbuG Q212 (CP001019), and CbuK Q154 (CP001020) were included. Each circle represents a unique genotype, and the size of the circle corresponds to the number of samples with that genotype. Branch labels and connecting lines correspond to the number of different markers between the genotypes. Genotypes connected by a gray background differ in only one marker from each other and may represent microvariants of one founder genotype.
Multiple different but apparently closely related MLVA genotypes, A to H, were identified in 33 clinical samples covering both acute Q fever patients (e.g., sputa, bronchoalveolar lavage [BAL] fluid, throat swabs) as well as chronic Q fever patients (e.g., heart valves, aorta tissue) (Table 1). A partial MLVA genotype (assigned as “p”) was obtained from another 13 samples that contained insufficient DNA to obtain a full profile. In all but one of the clinical samples that yielded a partial genotype, the same alleles were identified as those found in samples yielding a full genotype (Table 1). Clustering of the MLVA genotypes using the minimum spanning tree method showed a high degree of genetic similarity between the Dutch MLVA genotypes (Fig. 1B). Specifically, all but one of the obtained Dutch MLVA genotypes are interconnected by repeat number changes in one of the six markers (this involved either Ms23, Ms24, Ms27, and Ms34). One sample (Q056) yielded a genotype that differed in two markers from the other genotypes, and the alleles that were found in these two markers were also different from those observed in the other Dutch samples (Table 1). In contrast, the genotypes from five sequenced C. burnetii strains all differed in at least 3 markers from the Dutch genotypes. Negative control samples neither yielded a positive PCR result nor an MLVA result. The geographical distribution of the MLVA genotypes is shown in Fig. 1A. From the two genotypes that were observed most frequently (i.e., genotypes A and G), the G genotype apparently has spread across the entire affected area, whereas the distribution of genotype A appears to be restricted to the northeastern part of the affected region. The diversity indexes (D) of the individual markers for the Dutch population, calculated according to Simpson (7), were 0.48, 0.12, 0.06, 0.00, 0.06, and 0.31 for Ms23, Ms24, Ms27, Ms28, Ms33, and Ms34, respectively, versus 0.67, 0.79, 0.73, 0.74, 0.75, and 0.86, respectively, obtained from a reference collection of C. burnetii isolates from ticks, animal placenta, vaginal secretions, and milk and from human liver and blood (1). The D values observed in the Dutch population were much lower than in the reference population, indicating that the Dutch genotypes are much more closely related to each other and maybe arose from one founder genotype. The fact that multiple genotypes were obtained thus allows a certain degree of fine-structuring within the outbreak region.
Genotyping by using a 10-locus MLVA panel, including 4 out of the 6 markers used in our study, revealed one predominant MLVA genotype among goats and sheep throughout the affected Q fever area (5). In these samples, alleles were found in markers Ms24, Ms27, Ms28, and/or Ms34 that were identical to those in the human samples, implicating the goats and/or sheep as the most likely source of the outbreak. To substantiate this hypothesis, we included placenta samples from goats using all 6 markers from our MLVA panel. These goat samples were from 3 different locations in the outbreak area and contained the predominant ruminant C. burnetti genotype. MLVA genotype G was identified in all tested samples, illustrating the genotypic identity between C. burnetii from humans and goats. Compared to the 10-loci MLVA method, we chose the markers that seemed to be among the most discriminatory markers described (1), which enabled us to develop an easy-to-use and clearly distinguishable MLVA genotyping method.
Our results are consistent with a scenario where one MLVA genotype was introduced in the dairy animal population, where it is gradually diversifying over time while spreading over the country and being transmitted to humans. We believe that the sudden increase of Q fever infections in humans could have been facilitated by the expansion of intensive goat farming in the southeast of The Netherlands in the last 2 decades (4). Another possibility is that the Dutch C. burnetii isolates are from a hypervirulent lineage that may be disseminating more rapidly than other genotypes. Both explanations, however, require further investigation.
In conclusion, this study shows that the unprecedented and ongoing Q fever outbreak in The Netherlands involved not only multiple different but closely related MLVA genotypes found at several locations spread across the entire affected area during the Q fever outbreak years of 2007 to 2010, indicating a clonal spread of C. burnetii across The Netherlands.
Footnotes
Published ahead of print 21 December 2011
REFERENCES
- 1. Arricau-Bouvery N, et al. 2006. Molecular characterization of Coxiella burnetii isolates by infrequent restriction site-PCR and MLVA typing. BMC Microbiol. 6:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Karagiannis I, et al. 2009. Investigation of a Q fever outbreak in a rural area of The Netherlands. Epidemiol. Infect. 137:1283–1294 [DOI] [PubMed] [Google Scholar]
- 3. Klaassen CHW, Nabuurs-Franssen MH, Tilburg JJHC, Hamans MAWM, Horrevorts AM. 2009. Multigenotype Q fever outbreaks, the Netherlands. Emerg. Infect. Dis. 15:613–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Roest HIJ, et al. 2010. The Q fever epidemic in the Netherlands: history, onset, response and reflection. Epidemiol. Infect. 5:1–12 [DOI] [PubMed] [Google Scholar]
- 5. Roest HIJ, et al. 2011. Molecular epidemiology of Coxiella burnetii from ruminants in Q fever outbreak, the Netherlands. Emerg. Infect. Dis. 17:668–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schimmer B, et al. 2009. Sustained intensive transmission of Q fever in the south of the Netherlands, 2009. Euro Surveill. 14:19210. [DOI] [PubMed] [Google Scholar]
- 7. Simpson EH. 1949. Measurement of diversity. Nature 163:688 [Google Scholar]
- 8. Tilburg JJHC, et al. 2010. Interlaboratory evaluation of different extraction and real-time PCR methods for detection of Coxiella burnetii DNA in serum. J. Clin. Microbiol. 48:3923–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]