Abstract
Recently we evaluated a custom TaqMan array card (TAC) detection system, formerly known as a TaqMan low-density array (TLDA) card, for simultaneous real-time PCR identification of 21 pathogens and three control targets in duplicate from respiratory specimens (M. Kodani et al., J. Clin. Microbiol. 49:2175–2182, 2011). We engineered an adaptable and expandable system of in vitro RNA transcripts to serve as a combined positive control for both DNA and RNA targets in multiple-pathogen-detection technologies based on real-time reverse transcription-PCR.
TEXT
A positive-control template for singleplex real-time PCR can be easily obtained by purifying nucleic acids from a target organism. This is also feasible when a few targets are combined in the same multiplex reaction. However, in the case of multiple-pathogen-detection assays, such as the TaqMan array card (TAC) detection system, formerly known as TaqMan low-density arrays (TLDA), where 24 to 48 targets are amplified simultaneously from either RNA or DNA templates (6), obtaining a combined positive control (CPC) becomes more cumbersome. One major advantage of the TAC arrays is that total nucleic acids from one clinical specimen can be combined with PCR mix, loaded once into a TAC port, and separated by microfluidics into 48 separate reactions capable of amplifying either DNA or RNA (6). Thus, we attempted to engineer a positive control that can be loaded into one port on every card and used as a template for all assays in the panel.
The design of our synthetic combined positive control originated with a compilation of ideas used to make positive-control templates for various PCR assays, including using a series of genomic regions cloned into multiple plasmids as a multiplex control (7), synthetic oligonucleotide templates (10), and runoff RNA transcript templates (9). Some alternative positive-control designs utilized a separate fluorescent channel to monitor amplification efficiency and inhibition of PCR (2), but due to the nature of TAC cards, we were able to use the same channel and the exact primer and probe sequences to generate the positive control for all assays. The advantage of this CPC is its ability to monitor reagent quality, amplification efficiency, and inhibition, as well as monitoring homogeneity of assays over time and location. In addition, this design can be applied to both viral and bacterial targets. We evaluated our CPC design for flexibility to address any observed inconsistency in amplification, potential interference between oligonucleotide sets, theoretical RNA folding, specificity, and stability.
The first step in the design of the CPC system for real-time reverse transcription-PCR (rtRT-PCR) involved engineering a series of long synthetic oligonucleotides. For both DNA and RNA targets, we combined a forward primer sequence, followed by the probe sequence, and the reverse complement of the reverse primer sequence into an artificial template for each assay (see Fig. S1 in the supplemental material). The final construct was inserted into a plasmid. If the assay probe was originally designed to the reverse complement strand, the probe sequence was also reverse complemented on the plasmid. In addition, if two probes were required for the same assay, they were included in sequential order on the synthetic oligonucleotide, as long as they were designed to a different template region. When degenerate bases were used in an assay design, only one version of each primer or probe was engineered into the CPC plasmids. Because we attempted to develop a positive control for more than 50 targets, we decided to limit the size of each insert to around 1 kb, which allowed for inclusion of about 15 to 20 target assays per transcript. Each plasmid insert was designed to contain the T7 promoter sequence, the RNP3 human positive-control assay used on all TAC cards (6), 15 to 20 target assays, an Erwinia xeno assay for laboratory contamination control, and a reverse complement of the SP6 promoter sequence. One advantage of this design is that new assays can be added to the combined positive control by simply generating an additional plasmid containing all the elements listed above along with any new assay primer and probe sequences.
All plasmids and transcripts were handled in another laboratory to avoid potential contamination with templates for all assays commonly used in our laboratory. Upon receipt, lyophilized plasmids were handled according to the manufacturer's recommendations (IDT, Coralville, IA). A 1-in-1,000 dilution was made in water and used as a template in a PCR. The insert was amplified from each plasmid using M13 (−20) forward and M13 (−27) reverse primers available from IDT using 45 μl of Supermix (Invitrogen, Carlsbad, CA), 1 μl of each 10 μM primer, and 3 μl of the diluted plasmid template. The PCR cycling conditions were as follows: 94°C for 2 min, followed by 40 cycles of 94°C for 30 s, 50°C for 30 s, and 68°C for 90 s. The final extension was performed at 68°C for 10 min, followed by storage at 4°C. The size of each amplicon was checked by gel electrophoresis to ensure it matched the predicted size.
Five microliters of the amplicon from each plasmid was used as a template to generate the transcript using the SP6 MEGAscript kit according to the manufacturer's recommended protocol (Life Technologies, Carlsbad, CA). The reaction mixtures were incubated at 37°C for 2.5 h. The transcripts were subsequently purified using the MEGAclear kit as recommended by the manufacturer (Life Technologies, Carlsbad, CA). Following purification, all transcripts were initially diluted 1:100,000 in Tris-EDTA (TE) buffer (pH 8.0; Ambion, Austin, TX) spiked with yeast tRNA (50 ng/ml) and stored at −80°C in small aliquots.
The initial evaluation of transcripts was performed for each transcript in individual reverse transcription–real-time PCRs for RNP3 and Erwinia xeno as described previously (6). The Erwinia forward primer sequence was 5′ GCC GTT TGT TCA CGC TTT G 3′, the reverse primer was 5′ AGA GCC AGT CAG CCC GAT AG 3′, and the probe sequence was 5′ 6-carboxyfluorescein (FAM)-TGA ACG CCA GCA AGA CGA TCC AGT ATC-BHQ1 3′. Once the individual assay results were satisfactory, the transcripts were evaluated in respiratory panel TAC assays as described previously (6).
Individual transcripts were combined by diluting each to the 10−9 or 10−10 dilution in TE buffer (pH 8.0) (Ambion, Austin, TX) containing yeast tRNA (50 ng/ml). The combined positive control was aliquoted into single-use tubes and frozen at −80°C. Fourteen individual aliquots were thawed on ice and used on two respiratory panel TAC cards as described previously (6) to determine the extent of variability within a batch.
The sequence of each synthetic oligonucleotide was screened to ensure that primers and probes hybridized only to their specific locations on the transcript. This was accomplished using the Basic Local Alignment Tool (BLAST) in NCBI (1) by running the synthetic oligonucleotide sequence against the FASTA file of all primer and probe sequences used for our positive-control system to date. A few challenges became evident after this analysis. For instance, nonspecific binding of one oligonucleotide to a junction of a primer and a probe from another assay was observed. To remedy this problem, we added a short random nucleotide sequence spacer between the primer and the probe to reduce binding affinity and nonspecific amplification (see Fig. S1b in the supplemental material).
Although all synthetic oligonucleotides were designed to meet the above requirements, a couple of assays showed substandard performance upon testing by rtRT-PCR as judged by threshold cycle (CT) values that were higher than those of other assays and lower fluorescence levels (data not shown). Troubleshooting their performance led us to use the Mfold Web server for nucleic acid folding and hybridization prediction (11). Analysis of predicted RNA folds revealed theoretical stable hairpin structures present within the affected assays. This problem was resolved by placing primers and probes in a different configuration on the original synthetic oligonucleotide (see Fig. S1c in the supplemental material). During the reconfiguration, we kept the primers and probes in the correct orientation on the template to avoid having the PCR amplification fail. This conversion was performed with the Bordetella pertussis target 1 and RNP3 assays on a separate insert within pCP5 because the original transcript derived from pCP2a had a CT of 39.55 for B. pertussis target I. This was vastly improved to a CT of 29.48 following the conversion described for Fig. S1c. The CT values for the RNP3 assay were unchanged by this conversion. To demonstrate the specificity of the synthetic templates, individual transcripts as well as the pooled transcripts were run on the respiratory panel TAC card using the established protocols (6). Real-time amplification was observed only for the transcripts containing the appropriate assays (see Table S1 in the supplemental material).
The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) marker, used by Life Technologies (Carlsbad, CA) as a manufacturing quality control standard for all TAC cards, was not included on any of the synthetic oligonucleotides and as expected did not amplify in any of the ports on the card. This marker provided another layer of control, because a positive result in this well was suggestive of human nucleic acid contamination within the combined synthetic positive-control sample. Another human marker, RNP3, was included on every plasmid. When five transcripts were combined into a CPC in an equal molar ratio, the CT value for RNP3 dropped by 2.14 instead of the expected 1.65. This may have been due to steric hindrance or intermolecular interactions when five different transcripts and five times more RNA are present in the template solution. Ultimately, this observation had no deleterious effects on the CPC performance.
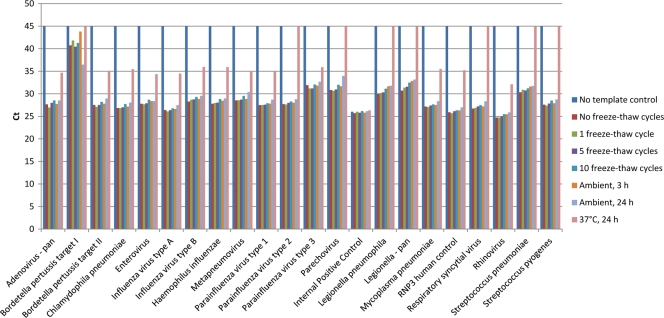
The stability of this genetically engineered CPC was tested by subjecting it to different freeze-thaw conditions and different temperatures for various lengths of time (Fig. 1). Up to 10 freeze-thaw cycles did not have a significant effect on the transcript performance on TAC cards. We noted a modest increase in the CT values at 10 freeze-thaw cycles; however, all assays still maintained a reliable qualitative answer (Fig. 1). In addition, we exposed the transcripts to different temperatures for various lengths of time. Although the CT values started increasing after 3 h of exposure to ambient temperature (21°C), qualitative results remained unchanged even after overnight exposure in this environment. Exposure to 37°C overnight resulted in a significant increase in CT values, with 8/22 (36.4%) assays displaying no amplification of the positive-control template. Collectively, these data suggested that positive-control transcripts were highly stable within normal operational parameters and that successful amplification could occur in spite of some mishandling. For global use, the CPC was prepared in yeast tRNA/TE buffer and stored as single-use aliquots.
Fig 1.
Stability of the combined-positive-control templates. The combined-positive-control templates were distributed into one-time-use aliquots, and different aliquots were exposed to various temperatures and numbers of freeze-thaw cycles. The CT values obtained on a respiratory panel TAC card were plotted per target and grouped by exposure to different experimental conditions. For better visualization of the results, all negative results were assigned a CT value of 45.
Reproducibility of the positive-control aliquots was demonstrated in Table 1. As a part of a larger task to generate 4,000 aliquots of the combined positive control for TAC arrays, a random sample of 14 tubes was tested on TAC cards. The average CT value, standard deviation, and range are reported for each assay (Table 1). Most assays performed in a tight range of 2 to 3 CT values. The Legionella—pan assay showed the highest range (8.24 CTs), but this may be attributed to the fact that one of 28 Legionella—pan assays amplified inefficiently. This occurrence may have resulted from a TAC malfunction during preparation or physical obstruction with the microfluidics channel. Upon exclusion of the one outlying value, the range for the pan-Legionella assay was within 1.6 CTs and the standard deviation dropped to 0.49. Additionally, the parechovirus assay showed a range of four CT values and a standard deviation of 1.21, suggesting less quantitative reproducibility than the majority of the assays. This observation may be attributed to the lower efficiency and fluorescence of this assay on the TAC card.
Table 1.
A survey of a random subset of 14 out of 4,000 single-use combined positive-control aliquots on the respiratory panel TAC card
| Target | CT |
|
|---|---|---|
| Avg (SD) | Range | |
| Influenza virus type A | 25.41 (0.55) | 24.75–26.51 |
| Influenza virus type B | 26.14 (0.50) | 25.28–27.08 |
| Respiratory syncytial virus | 27.67 (0.67) | 26.83–29.31 |
| Parainfluenza virus type 1 | 26.22 (0.86) | 24.58–27.55 |
| Parainfluenza virus type 2 | 26.44 (0.47) | 25.36–27.32 |
| Parainfluenza virus type 3 | 29.40 (0.78) | 28.40–31.15 |
| Metapneumovirus | 27.15 (0.79) | 26.07–28.61 |
| Rhinovirus | 24.35 (0.79) | 23.25–25.82 |
| Enterovirus | 26.96 (0.61) | 26.07–28.00 |
| Parechovirus | 29.06 (1.21) | 26.73–30.96 |
| Adenovirus—pan | 25.88 (0.56) | 25.15–26.98 |
| RNP3 human control | 25.70 (0.53) | 24.46–26.63 |
| Legionella—pan | 30.98 (1.48) | 29.90–38.14 |
| Haemophilus influenzae | 26.37 (0.86) | 24.97–27.53 |
| Streptococcus pneumoniae | 26.80 (0.44) | 26.08–27.67 |
| Streptococcus pyogenes | 25.89 (0.55) | 24.85–26.81 |
| Mycoplasma pneumoniae | 25.03 (0.54) | 24.41–26.17 |
| Chlamydophila pneumoniae | 24.14 (0.51) | 23.52–25.45 |
| Bordetella pertussis target I | 27.30 (0.57) | 26.18–28.64 |
| Bordetella pertussis target II | 26.11 (0.50) | 25.32–27.31 |
Overall, across all assays, there was a difference of approximately 2 logs between the lowest and the highest CT value even though the templates were present in an equal molar ratio. This effect may be due to differences in assay efficiencies and/or possible steric hindrance within and between RNA transcript molecules. We also examined the effect of assay location on the transcript on the CT value and found no correlation between the proximity to the end of the transcript and the CT value (data not shown).
In summary, we engineered a versatile positive-control system that can be used with multiple-pathogen-detection assays, as well as individual rtRT-PCR singleplex or multiplex assays. The process described here allows one to emulate the design procedure and application for customized assay targets. By using this approach, both RNA and DNA targets can be tested simultaneously, thus obviating the need for two separate preparations to account for each type of nucleic acid. Although this approach is restricted to reverse transcription-PCR chemistry, it affords the convenience of testing for both viral and bacterial targets on the same TAC. This is especially useful when testing for respiratory pathogens, since coinfections often exist (3, 4, 5, 8). Further, a single CPC preparation provides an opportunity to monitor TAC performance from card to card and allows normalization of the data. Primary design requires minimal modifications upon initial testing, with most difficulties being resolved by simply analyzing sequence comparisons and/or RNA folding predictions and making necessary adjustments to improve performance. This system is versatile because each assay can be independently subjected to troubleshooting at minimal cost. If only minor modifications are required to the original oligonucleotide, biotechnology companies can perform mutagenesis on the original plasmid, providing a significant cost savings over de novo synthesis. This system is relatively inexpensive, since one transcription reaction can generate enough positive control to last for thousands of TAC experiments and is easily expandable by creating additional transcripts and mixing them with the preexisting batch of the positive control. In addition, since we anticipated widespread use within our global partner laboratories, we determined adequate stability of the CPC over a wide range of temperatures and freeze-thaw conditions.
Large-scale multiple-pathogen-detection systems are creating a paradigm shift in the way infectious diseases are diagnosed and treated. These advancements come with new challenges, such as coordination and comparison of results across multiple platforms and multiple laboratories. The combined-positive-control system we have developed is a first step in overcoming some of these challenges.
Supplementary Material
ACKNOWLEDGMENTS
We thank Daniel Burken for assistance with bioinformatics for all plasmid insert sequences. We also thank Michael Bowen for use of isolated laboratory space and Dean Erdman and Brett Whitaker for helpful scientific discussions about plasmid insert design.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print 14 December 2011
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Auburn H, Zuckerman M, Broughton S, Greenough A, Smith M. 2011. Detection of nine respiratory RNA viruses using three multiplex RT-PCR assays incorporating a novel RNA internal control transcript. J. Virol. Methods 176:9–13 [DOI] [PubMed] [Google Scholar]
- 3. Jacoby P, et al. 2007. Modelling the co-occurrence of Streptococcus pneumoniae with other bacterial and viral pathogens in the upper respiratory tract. Vaccine 25:2458–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Johansson N, Kalin M, Tiveljung-Lindell A, Giske CG, Hedlund J. 2010. Etiology of community-acquired pneumonia: increased microbiological yield with new diagnostic methods. Clin. Infect. Dis. 50:202–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Johnstone J, Majumdar SR, Fox JD, Marrie TJ. 2008. Viral infection in adults hospitalized with community-acquired pneumonia: prevalence, pathogens, and presentation. Chest 134:1141–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kodani M, et al. 2011. Application of TaqMan low-density arrays for simultaneous detection of multiple respiratory pathogens. J. Clin. Microbiol. 49:2175–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lebo RV, Bixler M, Galehouse D. 2007. One multiplex control for 29 cystic fibrosis mutations. Genet. Test. 11:256–268 [DOI] [PubMed] [Google Scholar]
- 8. Oosterheert JJ, et al. 2005. Impact of rapid detection of viral and atypical bacterial pathogens by real-time polymerase chain reaction for patients with lower respiratory tract infection. Clin. Infect. Dis. 41:1438–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Snow M, McKay P, Matejusova I. 2009. Development of a widely applicable positive-control strategy to support detection of infectious salmon anaemia virus (ISAV) using Taqman real-time PCR. J. Fish Dis. 32:151–156 [DOI] [PubMed] [Google Scholar]
- 10. Whiley DM, et al. 2010. A simple approach for preparing real-time PCR positive reaction controls for rare or emerging viruses. J. Clin. Virol. 48:193–197 [DOI] [PubMed] [Google Scholar]
- 11. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.