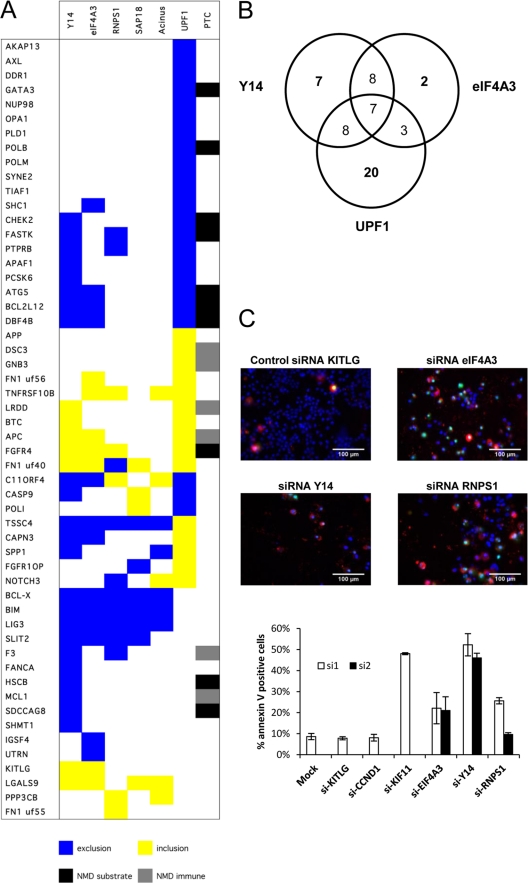
Fig 5.
EJC components control a network of splicing events linked to apoptosis. (A) RT-PCR analysis monitoring the alternative splicing of 54 units following the knockdown of EJC components and Upf1 in HeLa cells. Total RNA was extracted from HeLa cells at 72 h after transfection with siRNAs. The colored boxes represent significant shifts (minimum 10%) in the percent splicing index (PSI). The “PTC” column indicates the presence of a premature termination codon introduced by alternative splicing by either inclusion or exclusion. If the PTC is located more than 50 nucleotides upstream of an exon-exon junction, it is predicted to be a target for NMD and hence the event is indicated as a black box (NMD substrate). If the PTC is located within the 50 nucleotides upstream of the exon-exon junction or downstream of the last exonic junction, the transcript is assumed to escape NMD (NMD immune) and is indicated as a gray box. (B) Venn diagram showing the numbers of transcripts regulated by Y14, eIF4A3, and UPF1. Transcripts are considered coregulated by two or three of these proteins only if the splicing ratio shifts in the same direction upon depletion of the proteins. (C) Annexin V assay in HeLa cells transfected with siRNAs targeting EJC components. Cells were immunostained for annexin V (red), and nuclei were labeled with Hoechst stain (blue) (69). The percentage of apoptotic cells was quantitated (bottom graph) upon transfection of siRNAs targeting eIF4A3, Y14, and RNPS1 (2 siRNAs each) and compared to that for various controls (Lipofectamine alone, negative-control siRNAs targeting KITLG and cyclin D1, and a positive-control siRNA targeting KIF11 [71]).