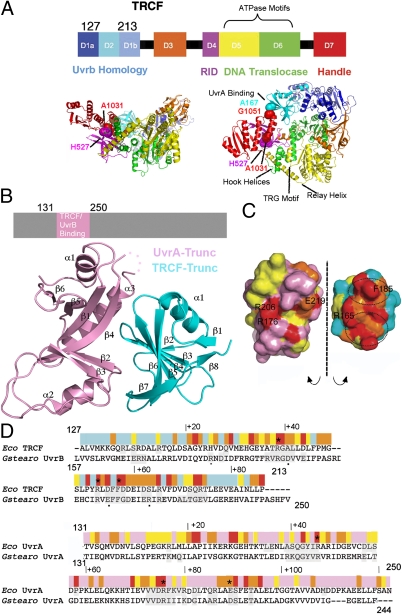
Fig. 1.
X-ray structure of the Escherichia coli TRCF–Trunc/UvrA–Trunc complex. (A) Structure of apo E. coli TRCF (PDB ID 2EYQ). Location of engineered Cys is indicated by spheres. (B) TRCF–UvrA core complex. (C) Solvent-accessible surface of the TRCF–UvrA complex, obtained by splaying the complex open and colored by evolutionary conservation as in D. (D) E. coli TRCF-Trunc/Geobacillus stearothermophilus UvrB sequence alignment with substitutions affecting UvrA binding (20, 27) marked by an asterisk in TRCF-Trunc and by a small black dot in UvrB. Sequence conservation is indicated with a color ramp from red (invariant) to cyan (variable). Interfacing residues in the TRCF–UvrA and UvrB–UvrA core complexes (20) are shaded in gray. The UvrA sequence is annotated similarly.