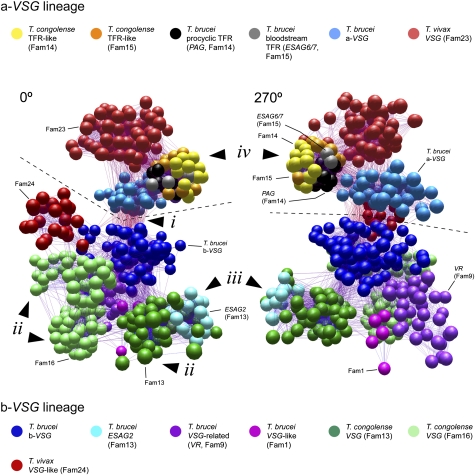
Fig. 1.
A sequence-similarity network of VSG-like sequences from African trypanosome genomes, shown from 0° and 270° angles. A 3D rendering of the network is provided as Movie S1. The network represents pairwise maximum likelihood protein distances, generated from multiple alignments of selected a-VSG–like (a-VSG, Fam23, TFR-like, and PAG-like proteins; n = 174) and b-VSG–like (b-VSG, Fam13, Fam16, Fam24, VR, ESAG2, and Fam1 proteins; n = 339) protein sequences, which are representative of global diversity. Spheres represent individual sequences shaded according to subfamily. A dashed line separates a-VSG–like subfamilies (above the line) and b-VSG subfamilies (below the line). Four significant features identified in the text are labeled: (i) sequence similarity between a-VSG and b-VSG lineages caused by the shared CTD of T. brucei VSG; (ii) diverse clusters of T. congolense b-VSG belonging to Fam13 and Fam16; (iii) the position of ESAG2 nested within Fam13; and (iv) tight cluster of TFR-like genes from both T. brucei and T. congolense.