Abstract
The cytochrome c oxidase Cox2 has been purified from native membranes of the hyperthermophilic eubacterium Aquifex aeolicus. It is a cytochrome ba3 oxidase belonging to the family B of the heme-copper containing terminal oxidases. It consists of three subunits, subunit I (CoxA2, 63.9 kDa), subunit II (CoxB2, 16.8 kDa), and an additional subunit IIa of 5.2 kDa. Surprisingly it is able to oxidize both reduced cytochrome c and ubiquinol in a cyanide sensitive manner. Cox2 is part of a respiratory chain supercomplex. This supercomplex contains the fully assembled cytochrome bc1 complex and Cox2. Although direct ubiquinol oxidation by Cox2 conserves less energy than ubiquinol oxidation by the cytochrome bc1 complex followed by cytochrome c oxidation by a cytochrome c oxidase, ubiquinol oxidation by Cox2 is of advantage when all ubiquinone would be completely reduced to ubiquinol, e.g., by the sulfide∶quinone oxidoreductase, because the cytochrome bc1 complex requires the presence of ubiquinone to function according to the Q-cycle mechanism. In the case that all ubiquinone has been reduced to ubiquinol its reoxidation by Cox2 will enable the cytochrome bc1 complex to resume working.
Keywords: cytochrome c oxidase, quinol oxidase, cyanide inhibition, protein complex interaction
The aerobic respiratory chain of mitochondria consists of several complexes named complex I–V (complex I, NADH∶quinone oxidoreductase; complex II, succinate∶quinone oxidoreductase; complex III, quinol∶cytochrome c oxidoreductase; complex IV, cytochrome c oxidase; complex V, ATP-synthase). Complexes I, III, and IV act in series and together they transfer electrons from NADH to molecular oxygen (dioxygen), whereas complex II provides a parallel entry for electrons into the respiratory chain. Complexes I, III, and IV directly couple the electron flow to proton translocation across the membrane (proton pumping) to enhance the electrochemical proton gradient which is used by the ATP-synthase to produce ATP (1). The composition of the aerobic respiratory chain of prokaryotes is more complex. Frequently prokaryotic respiratory chains are branched and many terminal oxidases, reducing dioxygen, can be found (2).
Most terminal oxidases belong to the heme-copper superfamily (HCO), which is divided into families A, B, and C (3). The superfamily is defined by the presence of a unique bimetallic active site, consisting of a high-spin heme (a heme A, O, or B, thus denoted a3, o3, or b3, respectively) and a closely associated copper atom (CuB) in subunit I. In the catalytic cycle dioxygen is bound to the heme iron and reduced to water. With a few exceptions heme-copper oxidases use either c-type cytochromes or quinols as electron donors. Cytochrome c oxidases of the A and B families possess two redox-active subunits. Subunit I contains an additional low-spin heme (A or B, and thus called a or b, respectively) and the bimetallic active site. Subunit II houses the CuA center consisting of two copper atoms (4, 5). CuA receives electrons from cytochrome c at the outer membrane surface and transfers them to the low-spin heme. Quinol oxidases share the conserved subunit I with cytochrome c oxidases, but their subunit II possesses neither a CuA center nor a cytochrome c binding site (6).
Respiratory chain supercomplexes, consisting of several neighboring complexes, have been detected in bacteria (7), fungi (8), and higher plant mitochondria (9), as well as in human mitochondria (10), and are widely accepted as allowing productive substrate channeling between respiratory chain components. The respiratory chain supercomplexes described so far with a terminal oxidase contain a cytochrome c oxidase, whereas quinol oxidases have not been reported to be part of any supercomplex. Structural models of supercomplexes isolated from mitochondria have been presented (11–13), but the functional analysis of respiratory chain supercomplexes is lagging behind. The cytochrome bc1 complex is one of the most common components present in the characterized supercomplexes. It contains three redox-active subunits: the Rieske protein with an iron-sulfur cluster; a cytochrome b containing two hemes b, and cytochrome c1 with a covalently bound heme c (14). The whole cytochrome bc1 complex catalyzes the two-electron oxidation of a quinol at the outer surface of the membrane and the transfer of one electron to cytochrome c, whereas the second electron is transferred back toward the inner surface of the membrane and used to reduce an oxidized quinone according to the Q-cycle mechanism (15).
With a maximal growth temperature of 95 °C (16) Aquifex aeolicus is one of the most thermophilic bacteria known. It is a chemolithoautotrophic bacterium growing under microaerobic conditions (17). Its genome has been sequenced and its size found to be only one third of that of Escherichia coli (17). For oxygen respiration, genes encoding a cytochrome bc1 complex, two cytochrome c oxidases (one belonging to family A, one to family B of the heme-copper oxidases), and the alternative cytochrome bd quinol oxidase (17) have been detected. The two cytochrome c oxidases are encoded by the genes coxA1 and coxA2 (subunits I), respectively, by coxB and coxB2 (subunits II), respectively, and subunit III by the gene coxC. The existence of a supercomplex consisting of at least the cytochrome bc1 complex and a cytochrome c oxidase has previously been suggested (18). A supercomplex containing a sulfide∶quinone oxidoreductase (SQR) in addition was isolated recently, but the functional connection between the cytochrome bc1 complex and the cytochrome c oxidase has not been characterized (19). SQR contributes to sulfide detoxification and it catalyzes quinone reduction taking the electrons from sulfide (20).
Here we present the purification of the B-family cytochrome c oxidase (Cox2) from native membranes of A. aeolicus. It is related to the cytochrome ba3 oxidase from Thermus thermophilus and consists of subunits I, II, and IIa. Quite surprisingly it shows a dual substrate specificity, being able to use both cytochrome c and quinols as electron donors. In the supercomplex it can switch between cytochrome c and ubiquinol as electron donors, ubiquinol appears to be used when present in amounts exceeding the capacity of the cytochrome bc1 complex. This property guarantees the fast removal of excess redox equivalents, which probably originate from toxic hydrogen sulfide. Regeneration of ubiquinone by Cox2 helps the cytochrome bc1 complex to work because this complex cannot operate when all ubiquinone has been converted to ubiquinol.
Results
Purification and Identification of Subunits by Mass Spectrometry and Sequence Corrections.
After solubilization by the mild detergent n-dodecyl-ß-d-maltoside the cytochrome c oxidase and the cytochrome bc1 complex were eluted as a single peak from ion-exchange chromatography and size-exclusion chromatography columns indicating that they form a supercomplex. All redox-active subunits of the cytochrome bc1 complex, namely cytochrome b (PetB), cytochrome c1 (Cyc), the Rieske protein (PetA), and the cytochrome c oxidase subunits I (CoxA2) and II (CoxB2) were detected in this supercomplex (Fig. S1A). In addition, an outer membrane protein C (OprC) was also found in our supercomplex preparation (Fig. S1A). Currently it is unclear whether OprC is a genuine component of the supercomplex function or whether it is a contamination. The pure cytochrome c oxidase (Cox2) could be isolated from the supercomplex by gel filtration after changing the detergent from n-dodecyl-ß-d-maltoside to ß-d-octylglucoside. Only the Cox2 subunits I and II could be detected as visible SDS-PAGE gel bands and identified by MALDI-MS (Fig. S1C, Table S1).
Cox2 subunit I is truncated at the C terminus (Fig. S2A) as indicated by the detection using MALDI-MS of tryptic peptides with masses of 1,245, 2,019, 2,035 Da, which could only be generated by trypsin from the C-terminally shortened subunit (Fig. S2B). The N terminus also seems to be three amino acids shorter than annotated, with the peptide MVW missing. Because of their overall hydrophobicity N-terminal peptides are not visible in the MS spectrum (Fig. S2B). The calculated average mass of the N- and C-terminally corrected CoxA2 is 63.9 kDa, which is observed by laser-induced liquid beam ion desorption mass spectrometry (LILBID-MS).
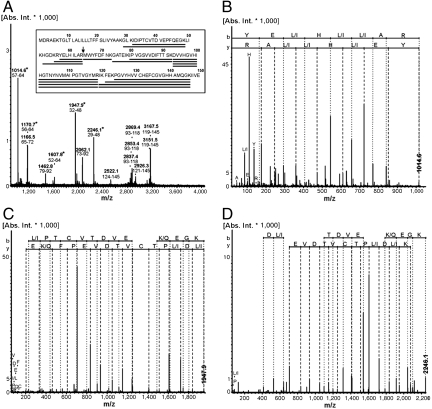
Several intense peaks in the MS spectra of Cox2 subunit II could not be assigned using the protein sequences provided by the databases (marked with asterisks in Fig. 1A). The spectrum of the fragmented peak at 1,014.6 Da was manually matched to the sequence YE(L/I)H(L/I)(L/I)AR (Fig. 1B). One possible peptide (YELHILAR) was found to be encoded in the 5′ region upstream of the annotated coxB2 gene by a blast search against the entire Aquifex genome (Fig. S3). By translating the region around the identified nucleotide sequence the correct CoxB2 sequence could be derived when the existence of a frame shift was considered (Fig. S3). This frameshift could be verified by de novo peptide sequencing using the MS/MS spectra of two further peaks at 1,947.9 and 2,246.1 Da (Fig. 1 C and D). The deduced peptide sequences IDIPTCVTDVEPFQEGK and GLKIDIPTCVTDVEPFQEGK covered the frameshift (Fig. S3). Because of the absence of peptides covering further sequence regions, the N terminus of the protein could not be unambiguously defined by MALDI-MS. DNA sequencing of a PCR product of coxB2 gene in this region confirmed the presence of the frameshift (Fig. S4). The corrected CoxB2 sequence is depicted in Fig. 1A (GenBank accession no. JN655694). CoxB2 has an average mass of 16.8 kDa, which fits exactly to the LILBID spectra.
Fig. 1.
Identification of a sequencing error for the deposited Cox2 subunit II (CoxB2). (A) MS spectrum of the trypsin digested Cox2 subunit II (band in Fig. S1C). Masses and sequence regions of assigned signals are labeled. Asterisks mark peaks verifying the new sequence regions. The bar plot aligns identified peptide mass fingerprint (PMF) fragments to the corresponding protein sequence regions. The arrow indicates the start of the incorrectly annotated Cox2 subunit II sequence; (B–D) MS/MS spectra of 1,014.6, 1,947.9, and 2,246.1. The manually interpreted peptide sequence is indicated (N-terminal b-ions, C-terminal y-ions, immonium ions). A differentiation between leucine/isoleucine (L/I) and lysine/glutamine (K/Q) is not possible. Because of the C-terminal specificity of the trypsin digest at lysine/arginine (K/R), the C terminus is solely indicated as K in C and D.
The subunit composition of the pure Cox2 and of the supercomplex was determined by LILBID-MS (Fig. S5). Cox2 consists of three subunits with masses of 5.2 kDa, 16.8 kDa, and 63.9 kDa. No cytochrome bc1 complex subunit was detected in this sample (Fig. S5A). The 63.9 kDa and the 16.8 kDa peaks were assigned to the truncated Cox2 subunit I and the corrected subunit II, respectively. A subunit with a mass of 5.1 kDa was detected in a supercomplex preparation containing Cox2 very recently (21). This 5.2 kDa subunit might be identical to this 5.1 kDa subunit. Its association with the Cox2 subunits I and II was evident by LILBID-MS (Fig. S5B). Cox2 has been proposed to belong to family B of the heme-copper terminal oxidases (3), and several members of this family [T. thermophilus ba3 (22), Natronobacterium pharaonis ba3 (23), Acidianus ambivalens aa3 (24), Sulfolobus acidocaldarius SoxABCD (25)] have been reported to possess a small subunit of about 50 amino acid residues. This subunit has been found by X-ray crystallography of the Thermus ba3 oxidase to take the position of the first transmembrane helix of subunit II in the family A cytochrome c oxidase from Parococcus denitrificans aa3, although in opposite orientation, and was therefore named subunit IIa (5). This feature can be expected to be the same in Aquifex Cox2 and in the Thermus ba3 oxidase. The 5.2 kDa subunit of the Aquifex Cox2 was therefore also named subunit IIa. Based on the subunit masses characterized by LILBID-MS and the expected 1∶1∶1 stoichiometry of the Cox2 subunits, the intact Cox2 has a mass of 85.9 kDa. A corresponding peak was detected by LILBID-MS (Fig. S5B), but an intact supercomplex could not be detected.
Characterization of the Prosthetic Groups.
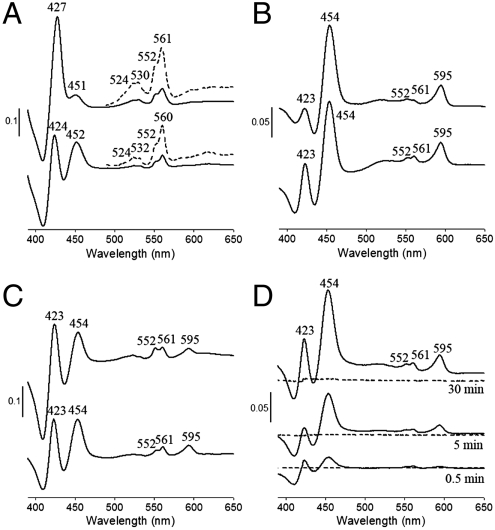
The prosthetic groups were characterized by UV-visible (Vis) absorption spectroscopy and EPR spectroscopy. The UV-Vis difference absorption spectra (dithionite reducing minus air oxidizing) of both Cox2 and the supercomplex showed typical peaks of heme b (532 nm, 560 nm in Cox2 and 530 nm, 560 nm in the supercomplex) and heme c (524 nm, 552 nm) (Fig. 2A). The Soret bands of these two hemes overlap (427 nm in the supercomplex). In addition, absorption peaks at 452 and 451 nm were detected in Cox2 and the supercomplex, respectively (Fig. 2A). In Thermus ba3 oxidase, the high-spin heme a3 has its Soret band at 442 nm (26, 27). The 452 nm absorption in Cox2 was also assigned to arise from a high-spin heme a3, but the corresponding alpha band (∼600 nm) was not detected in spectra of dithionite-reduced Cox2 and of the dithionite-reduced supercomplex. However, spectra of the pyridine hemochromes of extracted hemes from Cox2 and the supercomplex clearly show the absorption of the extracted heme A at 585 nm (Fig. S6). In addition, the observed 1.2∶1 ratio of hemes B and A in Cox2 based on the pyridine difference spectra shows that Cox2 is a cytochrome ba3 oxidase.
Fig. 2.
UV-Vis difference spectra of Cox2 and the supercomplex. The spectra were recorded at room temperature. (A) Dithionite reduced minus air oxidized Cox2 (lower) and supercomplex (upper). Dotted lines show the absorption signals with a magnification of 5× in the range of 490–600 nm. The spectra of reduced enzymes were recorded after 10 s when 200 mM dithionite were added into the cuvette. (B) decylubiquinol reduced minus air oxidized Cox2 (lower) and supercomplex (upper). Both spectra were recorded after proteins were incubated with 45 μM decylubiquinol in the presence/absence of 1 mM potassium cyanide for 30 min. (C) TMPD/ascorbate reduced minus air oxidized Cox2 (lower) and supercomplex (upper). The reducing spectra were recorded after 10 s when 0.3 mM TMPD/3 mM ascorbate and 1 mM potassium cyanide were applied. (D) Incubation of Cox2 with decylubiquinol and cyanide (solid lines) or cyanide alone (dotted lines). It shows that cyanide alone cannot induce the 595 nm absorption of Cox2 within 30 min.
The supercomplex and, surprisingly, Cox2 alone could also be reduced by decylubiquinol and N,N,N',N'-tetramethyl-ρ-phenylenediamine (TMPD)/ascorbate (Fig. 2 B and C) in the presence of cyanide. In addition to the typical bands of heme b and c, both the alpha band (595 nm) and Soret band (454 nm) of the heme a3 cyanide complex were visible (Fig. 2 B and C). The Thermus ba3 oxidase reacts with cyanide extremely slowly (28) when it is present in the oxidized form. In our case, no absorption band at 595 nm could be detected when the oxidized form of Cox2 was incubated with cyanide for 30 min, also indicating a very slow reaction with cyanide, whereas a significant absorption appeared immediately when ubiquinol was present (Fig. 2D). This result already indicates that Cox2 can be reduced by ubiquinol. Control experiments with the Thermus ba3 oxidase showed that ubiquinol does not accelerate the appearance of the 595 nm band in the presence of cyanide, indicating a fundamental difference between both enzymes. In the aa3 type oxidases the high-spin heme a3 mainly contributes to the Soret band whereas the low-spin heme a to the alpha band (29). The lack of a visible alpha band in spectra of dithionite reduced Cox2 indicates the presence of a high-spin heme a3 and provides further evidence that Cox2 is a cytochrome ba3 oxidase. The positions of the absorption peaks of the dithionite reduced hemes in the supercomplex differ slightly from those in the isolated Cox2 (Fig. 2A). However, upon reduction by ubiquinol or TMPD/ascorbate (Fig. 2 B and C), the positions are identical in the supercomplex and the isolated Cox2. This observation indicates that only Cox2 is reduced in the supercomplex by ubiquinol and TMPD/ascorbate.
The origin of the substantial heme c-like absorption in our Cox2 preparations remains a mystery. No staining of SDS-PAGE gel bands with 3,3′,5,5′-tetramethylbenzidine, which would detect covalently bound heme c (30), was observed (Fig. S7). In addition, neither subunit I nor subunit II contains the sequence signature CXXCH for binding heme c covalently, whereas subunit IIa (5.2 kDa) can be expected to be too small to bind heme c. The optical absorption spectra would argue for the presence of an additional heme c containing component in our Cox2 preparations which, however, could not be detected by mass spectrometry (LILBID or MALDI). To investigate the source of the heme c-like absorption, we purified Cox2 further by an additional preparative isoelectric focusing step. Cox2 yielded two bands with isoelectric points of 6.4 and 7.0, respectively. Both bands show the heme c-like absorption in an identical manner. This result supports the idea that the heme c-like absorption originates from Cox2 itself, and may be caused by a split absorption of heme b.
The EPR spectrum of Cox2 shows prominent signals from copper at g = 2.0 and from a high-spin heme a3 at g = 6.04 (Fig. S8A). The high-spin heme normally tightly couples with CuB leading to their invisibility in EPR spectra (31–33). However, because the high-spin heme a3 signal is visible in the EPR spectrum one would also expect to see that of CuB. The copper signal at g = 2.0 might therefore originate from both CuA and CuB. The sequence of the Aquifex Cox2 subunit II shows that all residues coordinating with CuA are conserved (Fig. S9). Signals from the low-spin heme b were detected at g = 3.4 and g = 3.0 in Cox2 (Fig. S8A), but the expected signal at g = 1.76 (19, 34) was not clearly visible in our Cox2 because of low resolution in this area. No low-spin hemes from the cytochrome bc1 complex were detected in our Cox2 preparation indicating complete separation of Cox2 and the cytochrome bc1 complex.
EPR signals from both Cox2 and the cytochrome bc1 complex were detected in the supercomplex. Besides copper and the high-spin heme a3, all corresponding peaks of the low-spin heme b of Cox2 were visible in the supercomplex (g = 3.4, g = 3.0, and g = 1.76 in Fig. S8B). Signals typical of the low-spin hemes from the cytochrome bc1 complex could be identified at g-values 3.7–3.3 (34). In addition the characteristic EPR spectrum of a Rieske iron-sulfur cluster can be detected in the supercomplex after reduction. The signal at g = 3.2 could be suggested to originate from the b-type cytochrome of hydrogenase I of A. aeolicus (34). However, the corresponding protein subunit was not detected in the supercomplex, and the g = 3.2 signal must remain unassigned.
Enzymatic Activities of Cox2 and the Supercomplex.
Surprisingly the isolated Cox2 oxidizes both cytochrome c and ubiquinol and uses their electrons to reduce molecular oxygen. The supercomplex oxidizes/reduces cytochrome c, and oxidizes ubiquinol (Table 1). The isolated Cox2 is able to oxidize horse heart cytochrome c (0.41 ± 0.01 U/mg) or uses the alternative electron donor TMPD/ascorbate to reduce oxygen. It was reported that Thermus ba3 oxidase subunit II lacks the acidic residues which contribute to the electrostatic interactions with horse heart cytochrome c whereas Paracoccus aa3 oxidase possesses these acidic residues; alternatively, it uses cytochrome c552 from the same organism (5, 35). Sequence alignment of CoxB2 with Paracoccus aa3 oxidase subunit II shows that most of the negatively charged residues belonging to the potential cytochrome c binding site are present in Aquifex Cox2 (Fig. S9). This observation supports the possibility that Aquifex Cox2 reacts with horse heart cytochrome c. This finding indicates that the interaction of Cox2 with its electron donating cytochrome c is different from that of the Thermus ba3 oxidase. Cox2 also oxidizes ubiquinol and the reaction rate is similar to that of the supercomplex, which indicates that quinol oxidation by the supercomplex may be catalyzed by Cox2 alone (Table 1). This assumption is in agreement with the UV-Vis absorption spectra (only Cox2 could be reduced by ubiquinol in the supercomplex, Fig. 2B). The isolated Cox2 is also able to act as a quinol∶cytochrome c oxidoreductase when cyanide is present (Table 1). This reaction is specifically described in Results.
Table 1.
Activities of Cox2 and the supercomplex
| Cytochrome c oxidation | Cytochrome c reduction in the presence of cyanide | Ubiquinol oxidation | |
| Cox2 | 0.41 ± 0.01 | 0.18 ± 0.04 | 1.22 ± 0.13 |
| Supercomplex | 1.16 ± 0.12 | 2.12 ± 0.34 | 0.96 ± 0.23 |
The enzymatic reactions were measured spectrophotometrically at 80 °C. Specific activities are shown by mean ± standard error from three measurements. The unit of micromole per minute per milligram was applied.
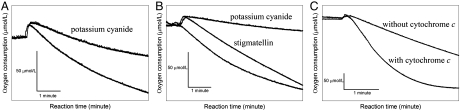
The reduction of oxygen in the presence of decylubiquinol was measured using an oxygen electrode (Fig. 3). Cox2 is able to reduce oxygen with decylubiquinol as an electron donor, and the reaction is inhibited by potassium cyanide (Fig. 3A). Likewise, the supercomplex reduces oxygen using decylubiquinol as electron donor. This reaction is inhibited by potassium cyanide (Fig. 3B), but almost unaffected by stigmatellin (Fig. 3B). Addition of oxidized cytochrome c to the supercomplex substantially accelerates the rate of oxygen reduction (Fig. 3C). The insensitivity to stigmatellin of the ubiquinol oxidation activity and its inhibition by cyanide clearly demonstrate that it is Cox2 rather than the cytochrome bc1 complex that is responsible for the oxidation of ubiquinol in the supercomplex. The acceleration of the ubiquinol oxidation activity of the supercomplex by added cytochrome c indicates that the cytochrome bc1 complex in the supercomplex becomes active upon addition of its substrate cytochrome c (the electron acceptor of the cytochrome bc1 complex) and also demonstrates the absence of a functional link (e.g., a membrane anchored electron shuttling cytochrome c) between the cytochrome bc1 complex and Cox2 in the supercomplex.
Fig. 3.
Oxygen consumption by Cox2 and the supercomplex in the presence of decylubiquinol and DTT. Reactions were recorded using an oxygen electrode at 40 °C. (A) Oxygen consumption by Cox2. The reaction was inhibited by 1 mM potassium cyanide. (B) Oxygen consumption by the supercomplex. This reaction was not inhibited by 40 μM stigmatellin, but inhibited by 1 mM potassium cyanide. (C) Oxygen consumption by the supercomplex. This reaction could be accelerated by addition of air oxidized horse heart cytochrome c (60 μM). The cytochrome c has to be injected together with the protein to avoid direct reduction by DTT. The scale bars indicate a change in the dioxygen concentration by micromole per liter per minute.
Ubiquinol Titration of the Supercomplex.
As shown above Cox2 oxidizes both cytochrome c and ubiquinol. We have investigated this reaction further to obtain more information about the role of the two-electron donors. Decylubiquinol was added to concentrations from 30 to 105 μM to investigate ubiquinol oxidation by the supercomplex in the presence or absence of cytochrome c and potassium cyanide as a Cox2 inhibitor (Fig. 4). The reaction rate of ubiquinol oxidation by Cox2 increases up to ubiquinol concentrations of 60 μM, above which it stays constant (Fig. 4A). This reaction is inhibited by cyanide. In the supercomplex the cytochrome bc1 complex oxidizes ubiquinol and produces reduced cytochrome c, which accumulates because of the presence of cyanide. The rates of cytochrome c reduction and ubiquinol oxidation both increase up to ubiquinol concentrations of 60 μM but are inhibited at higher ubiquinol concentrations (Fig. 4B). The observation that both cytochrome c reduction and ubiquinol oxidation are lower at high ubiquinol concentrations might be explained by the fact that oxidized ubiquinol (ubiquinone) is also a substrate of the cytochrome bc1 complex in the Q-cycle mechanism, the reaction suffering from the competition of ubiquinone by ubiquinol at high ubiquinol concentrations in a kind of substrate inhibition. When both Cox2 and the cytochrome bc1 complex are active in the supercomplex (Fig. 4C), the observed net cytochrome c reduction is slightly lower than that catalyzed by the cytochrome bc1 complex alone. This observation is because of the fact that Cox2 reoxidizes the reduced cytochrome c. At ubiquinol concentrations lower than 60 μM, the ubiquinol oxidation rate is similar to that catalyzed by the cytochrome bc1 complex (Fig. 4B), which indicates that ubiquinol is oxidized primarily by the cytochrome bc1 complex and that Cox2 then uses cytochrome c as an electron donor. At ubiquinol concentrations higher than 60 μM, the rate of ubiquinol oxidation remains nearly constant and similar to that catalyzed by Cox2 (Fig. 4A) alone, which is much higher than that catalyzed by the cytochrome bc1 complex (Fig. 4B). This observation indicates that Cox2 uses ubiquinol as electron donor when ubiquinol is present in excess. A careful determination of the KM values for ubiquinol of both the cytochrome bc1 complex and Cox2 will shed further light into the relative activities of both complexes.
Fig. 4.
The effect of increasing decylubiquinol concentrations on the supercomplex activities. All reactions were measured spectrophotometrically at 80 °C. (A) Decylubiquinol oxidation in the presence (dashed line) and absence (solid line) of 1 mM potassium cyanide. (B) Cytochrome c reduction (dashed line) and decylubiquinol oxidation (solid line) in the presence of 60 μM horse heart cytochrome c and 1 mM potassium cyanide. (C) Cytochrome c reduction (dashed line) and decylubiquinol oxidation (solid line) in the presence of 60 μM horse heart cytochrome c. Error bars indicate the standard errors of the mean from three experiments. The y axis shows the enzyme-specific activity in percent.
Cyanide Poisoned Cox2 Can Act as a Ubiquinol∶Cytochrome c Oxidoreductase.
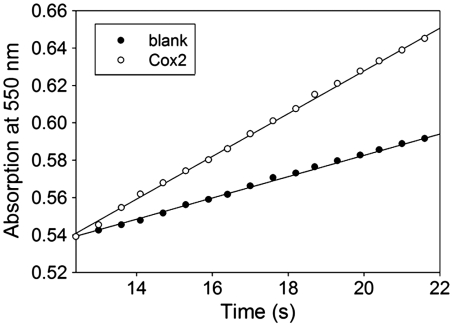
If Cox2 oxidizes both ubiquinol and reduced cytochrome c it might be able to transfer electrons from ubiquinol to oxidized cytochrome c under anaerobic conditions or when the binuclear active site is blocked. In this case electrons might flow from ubiquinol to the low-spin heme b and then back to CuA and oxidized cytochrome c when further transfer to the binuclear site and onto dioxygen and the intermediates of the catalytic cycle is not possible. Fig. 5 shows that this reaction is indeed the case. However, the speed of this artificial reaction is considerably lower than that of the same reaction catalyzed by the cytochrome bc1 complex. Such a lower activity has to be expected because oxidized cytochrome c is a product and not a substrate of the natural reaction of Cox2, and a prolonged binding of oxidized cytochrome c would lead to substrate inhibition.
Fig. 5.
Cytochrome c reduction of Cox2 in the presence of 1 mM potassium cyanide and 45 μM decylubiquinol. The reaction was measured spectrophotometrically at 80 °C as described in SI Materials and Methods for cytochrome c reduction. A specific activity of 0.53 μmol/ min per milligram was calculated for the Cox2 ubiquinol∶cytochrome c oxidoreductase activity. For the blank measurements buffer without Cox2 was used.
Discussion
As outlined in the results section Aquifex Cox2 is a cytochrome ba3 oxidase. Like other heme-copper terminal oxidases of the B family it consists of subunits I (CoxA2), II (CoxB2), and IIa. Our data show convincingly that it oxidizes both cytochrome c and ubiquinol and is therefore a cytochrome c oxidase and a quinol oxidase at the same time. This property is unique although cytochrome c oxidases and quinol oxidases belong to the same enzyme families (36) characterized by the presence of a homologous catalytic subunit I (37). Already the observation that air oxidized Cox2 reacts with cyanide in the presence of ubiquinol but not in its absence indicates that ubiquinol is able to reduce Cox2. It is unclear whether ubiquinol provides its electrons to heme b or CuA, but the hydrophobicity of decylubiquinol is a unique argument in favor of a direct reduction of heme b. The result, that cyanide inhibited Cox2 can act as a ubiquinol∶cytochrome c oxidoreductase provides evidence for the existence of different binding sites for cytochrome c and ubiquinol. However, kinetic experiments will still be required to determine whether heme b or CuA is the immediate electron acceptor for electrons from ubiquinol. Such kinetic experiments also may shed light onto the nature of the mysterious heme c type absorption in the visible absorption spectrum of reduced Cox2.
The control experiments with the Thermus ba3 oxidase demonstrate that a dual substrate specificity is not a general property of the heme-copper terminal oxidases of the B family, but possibly an invention of A. aeolicus. This invention must have provided an evolutionary advantage despite the fact that electron transfer from ubiquinol to oxygen via the cytochrome bc1 complex and a cytochrome c oxidase conserves more energy than via a quinol oxidase. Having in mind that the B family terminal oxidases appear to pump only two protons per cycle (38, 39) a direct quinol oxidation by a quinol oxidase is only half as energy efficient as the oxidation via the cytochrome bc1 complex and Cox2. There must be a compelling advantage for the usage of quinol oxidation by Cox2, in particular because the cytochrome bc1 complex and Cox2 are constituents of the same supercomplex.
The ubiquinol titration experiment shows that at high ubiquinol concentrations the ubiquinol oxidation by the supercomplex is much higher than that catalyzed by the cytochrome bc1 complex. Because the ubiquinol oxidation by cytochrome bc1 complex leads to cytochrome c reduction in the supercomplex, ubiquinol oxidation catalyzed by the cytochrome bc1 complex can be calculated from the observed amount of reduced cytochrome c. The difference of ubiquinol oxidation by the cytochrome bc1 complex and by the supercomplex is therefore caused by Cox2. The difference shows that in the supercomplex Cox2 uses cytochrome c as an electron donor at low ubiquinol concentration when cytochrome bc1 complex is active, but uses ubiquinol as an electron donor at high ubiquinol concentrations. High ubiquinol concentrations can be expected to be inhibitory upon the activity of the cytochrome bc1 complex, because in the cytochrome bc1 complex ubiquinone, the product of ubiquinol oxidation at the Qo-site, is also a substrate at the Qi-site according to the Q-cycle mechanism (15). At high ubiquinol to ubiquinone ratios ubiquinol will compete with ubiquinone at the Qi-site leading to inhibition. In the complete absence of ubiquinone the cytochrome bc1 complex cannot operate at all. There are a number of quinone reducing enzymes in A. aeolicus. Apart of the respiratory complex I a sulfide∶quinone oxidoreductase is present in vast amounts so that it could be crystallized and its structure could be determined (40). A sudden burst in sulfide in the natural environment of A. aeolicus may lead to a complete reduction of the ubiquinone pool and as a result the cytochrome bc1 complex would become inactive. As a consequence even in the presence of oxygen ubiquinol cannot be oxidized by the cytochrome bc1 complex. Only a quinol oxidase could remedy the situation, and this requirement might be the explanation why Aquifex Cox2 is able to oxidize ubiquinol. The Aquifex genome also encodes a quinol oxidase of the bd type. This enzyme might not be sufficiently active or unsuited for different reasons. The location of the ubiquinol oxidating Cox2 in the same supercomplex as the cytochrome bc1 complex certainly would constitute an advantage for rescuing the activity of the cytochrome bc1 complex by Cox2 over a recovery by the bd oxidase.
Materials and Methods
Membranes from A. aeolicus were prepared as described previously (41). Detailed procedures for protein isolation, MALDI- and LILBID-MS identification, and EPR spectroscopy, activity measurements are described in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Tanja Hedderich for excellent technical assistance, the Archaeenzentrum (Regensburg, Germany) for supplying the A. aeolicus cells, Tewfik Soulimane and Mohamed Noor for providing the Thermus ba3 oxidase, Julian Langer for performing additional mass spectrometric control experiments, Janet Vonck and Deryck Mills for performing additional single particle electron microscopic control experiments and Ulrich Brandt for providing the EPR facilities. We are grateful to David Parcej for reading the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft (SFB 472, SFB 628), the Max Planck Gesellschaft, and the Cluster of Excellence Macromolecular Complexes (Frankfurt, Germany).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database, www.ncbi.nlm.nih.gov/genbank (accession no. JN655694).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1121040109/-/DCSupplemental.
References
- 1.Saraste M. Oxidative phosphorylation at the fin de siecle. Science. 1999;283:1488–1493. doi: 10.1126/science.283.5407.1488. [DOI] [PubMed] [Google Scholar]
- 2.Thony-Meyer L. Biogenesis of respiratory cytochromes in bacteria. Microbiol Mol Biol Rev. 1997;61:337–376. doi: 10.1128/mmbr.61.3.337-376.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pereira MM, Santana M, Teixeira M. A novel scenario for the evolution of haem-copper oxygen reductases. Biochim Biophys Acta. 2001;1505:185–208. doi: 10.1016/s0005-2728(01)00169-4. [DOI] [PubMed] [Google Scholar]
- 4.Iwata S, Ostermeier C, Ludwig B, Michel H. Structure at 2.8 Å resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 5.Soulimane T, et al. Structure and mechanism of the aberrant ba(3)-cytochrome c oxidase from Thermus thermophilus. EMBO J. 2000;19:1766–1776. doi: 10.1093/emboj/19.8.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abramson J, et al. The structure of the ubiquinol oxidase from Escherichia coli and its ubiquinone binding site. Nat Struct Biol. 2000;7:910–917. doi: 10.1038/82824. [DOI] [PubMed] [Google Scholar]
- 7.Berry EA, Trumpower BL. Isolation of ubiquinol oxidase from Paracoccus denitrificans and resolution into cytochrome bc1 and cytochrome c-aa3 complexes. J Biol Chem. 1985;260:2458–2467. [PubMed] [Google Scholar]
- 8.Krause F. Detection and analysis of protein-protein interactions in organellar and prokaryotic proteomes by native gel electrophoresis: (Membrane) protein complexes and supercomplexes. Electrophoresis. 2006;27:2759–2781. doi: 10.1002/elps.200600049. [DOI] [PubMed] [Google Scholar]
- 9.Krause F, et al. Respirasome-like supercomplexes in green leaf mitochondria of spinach. J Biol Chem. 2004;279:48369–48375. doi: 10.1074/jbc.M406085200. [DOI] [PubMed] [Google Scholar]
- 10.Schagger H, et al. Significance of respirasomes for the assembly/stability of human respiratory chain complex I. J Biol Chem. 2004;279:36349–36353. doi: 10.1074/jbc.M404033200. [DOI] [PubMed] [Google Scholar]
- 11.Schafer E, Dencher NA, Vonck J, Parcej DN. Three-dimensional structure of the respiratory chain supercomplex I1III2IV1 from bovine heart mitochondria. Biochemistry. 2007;46:12579–12585. doi: 10.1021/bi700983h. [DOI] [PubMed] [Google Scholar]
- 12.Heinemeyer J, Braun HP, Boekema EJ, Kouril R. A structural model of the cytochrome C reductase/oxidase supercomplex from yeast mitochondria. J Biol Chem. 2007;282:12240–12248. doi: 10.1074/jbc.M610545200. [DOI] [PubMed] [Google Scholar]
- 13.Schafer E, et al. Architecture of active mammalian respiratory chain supercomplexes. J Biol Chem. 2006;281:15370–15375. doi: 10.1074/jbc.M513525200. [DOI] [PubMed] [Google Scholar]
- 14.Berry EA, Guergova-Kuras M, Huang LS, Crofts AR. Structure and function of cytochrome bc complexes. Annu Rev Biochem. 2000;69:1005–1075. doi: 10.1146/annurev.biochem.69.1.1005. [DOI] [PubMed] [Google Scholar]
- 15.Mitchell P. Possible molecular mechanisms of the protonmotive function of cytochrome systems. J Theor Biol. 1976;62:327–367. doi: 10.1016/0022-5193(76)90124-7. [DOI] [PubMed] [Google Scholar]
- 16.Huber R, Eder W. Aquificales. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. The Prokaryotes: A Handbook on the Biology of Bacteria: Proteobacteria: Delta and Epsilon Subclasses. Deeply Rooting Bacteria. 3rd Ed. Vol 7. New York: Springer; 2006. pp. 925–938. [Google Scholar]
- 17.Deckert G, et al. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 18.Guiral M, et al. New insights into the respiratory chains of the chemolithoautotrophic and hyperthermophilic bacterium Aquifex aeolicus. J Proteome Res. 2009;8:1717–1730. doi: 10.1021/pr8007946. [DOI] [PubMed] [Google Scholar]
- 19.Prunetti L, et al. New functional sulfide oxidase-oxygen reductase supercomplex in the membrane of the hyperthermophilic bacterium Aquifex aeolicus. J Biol Chem. 2010;285:41815–41826. doi: 10.1074/jbc.M110.167841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marcia M, et al. Characterizing a monotopic membrane enzyme. Biochemical, enzymatic and crystallization studies on Aquifex aeolicus sulfide∶quinone oxidoreductase. Biochim Biophys Acta. 2010;1798:2114–2123. doi: 10.1016/j.bbamem.2010.07.033. [DOI] [PubMed] [Google Scholar]
- 21.Prunetti L, Brugna M, Lebrun R, Giudici-Orticoni MT, Guiral M. The elusive third subunit IIa of the bacterial B-type oxidases: The enzyme from the hyperthermophile Aquifex aeolicus. PLoS One. 2011;6:e21616. doi: 10.1371/journal.pone.0021616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soulimane T, Than ME, Dewor M, Huber R, Buse G. Primary structure of a novel subunit in ba3-cytochrome oxidase from Thermus thermophilus. Protein Sci. 2000;9:2068–2073. doi: 10.1110/ps.9.11.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mattar S, Engelhard M. Cytochrome ba3 from Natronobacterium pharaonis—an archaeal four-subunit cytochrome-c-type oxidase. Eur J Biochem. 1997;250:332–341. doi: 10.1111/j.1432-1033.1997.0332a.x. [DOI] [PubMed] [Google Scholar]
- 24.Purschke WG, Schmidt CL, Petersen A, Schafer G. The terminal quinol oxidase of the hyperthermophilic archaeon Acidianus ambivalens exhibits a novel subunit structure and gene organization. J Bacteriol. 1997;179:1344–1353. doi: 10.1128/jb.179.4.1344-1353.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lubben M, Kolmerer B, Saraste M. An archaebacterial terminal oxidase combines core structures of two mitochondrial respiratory complexes. EMBO J. 1992;11:805–812. doi: 10.1002/j.1460-2075.1992.tb05117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerscher S, Hildebrandt P, Buse G, Soulimane T. The active site structure of ba3 oxidase from Thermus thermophilus studied by resonance raman spectroscopy. Biospectroscopy. 1999;5(Suppl 5):S53–S63. doi: 10.1002/(SICI)1520-6343(1999)5:5+<S53::AID-BSPY6>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 27.Zimmermann BH, Nitsche CI, Fee JA, Rusnak F, Munck E. Properties of a copper-containing cytochrome ba3: A second terminal oxidase from the extreme thermophile Thermus thermophilus. Proc Natl Acad Sci USA. 1988;85:5779–5783. doi: 10.1073/pnas.85.16.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Surerus KK, et al. Reaction of cyanide with cytochrome ba3 from Thermus thermophilus: Spectroscopic characterization of the Fe(II)a3-CN.Cu(II)B-CN complex suggests four 14N atoms are coordinated to CuB. Proc Natl Acad Sci USA. 1992;89:3195–3199. doi: 10.1073/pnas.89.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Antalis TM, Palmer G. Kinetic characterization of the interaction between cytochrome oxidase and cytochrome c. J Biol Chem. 1982;257:6194–6206. [PubMed] [Google Scholar]
- 30.Thomas PE, Ryan D, Levin W. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal Biochem. 1976;75:168–176. doi: 10.1016/0003-2697(76)90067-1. [DOI] [PubMed] [Google Scholar]
- 31.Falk KE, Vanngard T, Angstrom J. Heme spin-states of cytochrome c oxidase derived from room temperature magnetic susceptibility measurements. FEBS Lett. 1977;75:23–27. doi: 10.1016/0014-5793(77)80044-6. [DOI] [PubMed] [Google Scholar]
- 32.Thomson AJ, Brittain T, Greenwood C, Springall JP. Variable-temperature magnetic-circular-dichroism spectra of cytochrome c oxidase and its derivatives. Biochem J. 1977;165:327–336. doi: 10.1042/bj1650327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tweedle MF, Wilson LJ. Electronic state of heme in cytochrome oxidase III. The magnetic susceptibility of beef heart cytochrome oxidase and some of its derivatives from 7–200 K. Direct evidence for an antiferromagnetically coupled Fe (III)/Cu (II) pair. J Biol Chem. 1978;253:8065–8071. [PubMed] [Google Scholar]
- 34.Schutz M, et al. The naphthoquinol oxidizing cytochrome bc1 complex of the hyperthermophilic knallgasbacterium Aquifex aeolicus: Properties and phylogenetic relationships. Biochemistry. 2003;42:10800–10808. doi: 10.1021/bi034452a. [DOI] [PubMed] [Google Scholar]
- 35.Maneg O, Malatesta F, Ludwig B, Drosou V. Interaction of cytochrome c with cytochrome oxidase: Two different docking scenarios. Biochim Biophys Acta. 2004;1655:274–281. doi: 10.1016/j.bbabio.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 36.Musser SM, Stowell MH, Chan SI. Comparison of ubiquinol and cytochrome c terminal oxidases. An alternative view. FEBS Lett. 1993;327:131–136. doi: 10.1016/0014-5793(93)80156-o. [DOI] [PubMed] [Google Scholar]
- 37.Calhoun MW, Thomas JW, Gennis RB. The cytochrome oxidase superfamily of redox-driven proton pumps. Trends Biochem Sci. 1994;19:325–330. doi: 10.1016/0968-0004(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 38.Han H, et al. Adaptation of aerobic respiration to low O2 environments. Proc Natl Acad Sci USA. 2011;108:14109–14114. doi: 10.1073/pnas.1018958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kannt A, et al. Electrical current generation and proton pumping catalyzed by the ba3-type cytochrome c oxidase from Thermus thermophilus. FEBS Lett. 1998;434:17–22. doi: 10.1016/s0014-5793(98)00942-9. [DOI] [PubMed] [Google Scholar]
- 40.Marcia M, Ermler U, Peng G, Michel H. The structure of Aquifex aeolicus sulfide∶quinone oxidoreductase, a basis to understand sulfide detoxification and respiration. Proc Natl Acad Sci USA. 2009;106:9625–9630. doi: 10.1073/pnas.0904165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng G, et al. Isolation, characterization and electron microscopic single particle analysis of the NADH∶ubiquinone oxidoreductase (complex I) from the hyperthermophilic eubacterium Aquifex aeolicus. Biochemistry. 2003;42:3032–3039. doi: 10.1021/bi026876v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.