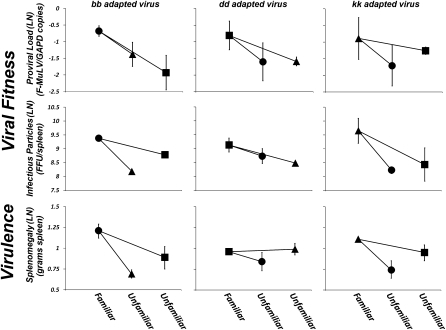
Fig. 4.
Pathogens adapt to specific MHC genotypes, and this results in lower fitness and virulence when infecting unfamiliar MHC genotypes. Pathogen adaptation to specific MHC genotypes is predicted to result in strong host genotype by pathogen genotype interactions. To test this prediction six groups of animals (6–10 individuals per group) from each of our three host genotypes were independently infected with all six postpassage stocks (two passage lines were conducted in each of the three host genotypes) and interaction effects were tested using a least-square general linearized model. Symbols reflect host genotypes (●, BALB/cbb hosts; ■, BALB/cdd hosts; ▲, BALB/ckk hosts) and represent the least-square mean of two replicate tests (error bars represent the range of means for each test). Data are shown on a natural log scale. Comparisons of patterns of pathogen adaptation and virulence between virus stocks (denoted bb passaged virus, dd passaged virus, and kk passaged virus) across the three host genotypes shows that pathogen fitness and virulence is host genotype-specific. Viral fitness (12 of 12 comparisons) and disease virulence (5 of 6 comparisons) are higher when a passaged virus stock is exposed to a familiar (i.e., its’ host genotype-of-passage) versus an unfamiliar (a genotype it hasn't been passaged through) MHC genotype. On average (based on least-squares means shown above), viral fitness is 114% (x = −0.79 F-MuLV/GAPDH ratio versus x = −1.56 F-MuLV/GAPDH ratio) and 149% (x = 9.41 FFU/spleen vs. x = 8.50 FFU/spleen) higher in familiar vs. unfamiliar host genotypes based on proviral load and infectious particles, respectively. Virulence is 28% (x = 1.10 g spleen vs. x = 0.86 g spleen) higher in familiar vs. unfamiliar host genotypes.