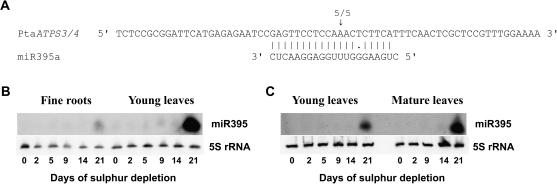
Fig. 5.
(A) Validation of PtaATPS3/4 as the target of miR395. Validation was carried out by rapid amplification of 5′ cDNA ends (5′ RACE). RNA from mature leaves after 0 d or 21 d of sulphur depletion treatment and a primer pair for PtaATPS3/4 products were used (see materials and methods). Each sample derived from two plants pooled together. Arrows indicate the 5′ ends of cleavage products mapped inside the displayed sequence including the number of cloned and sequenced RACE products with the respective 5′ end. Target sequences are shown on top of the miR395a sequence. (B, C) Expression of miR395a in fine roots, young leaves, and mature leaves of poplar due to sulphur depletion treatment. Northern blot analysis was carried out with 15 μg of total RNA from fine roots and young leaves using a high-sensitive membrane (Amersham-Hybond-NX) (B) or with 70 μg of total RNA from young leaves and mature leaves using a less sensitive membrane (Amersham-Hybond-N+) (C). All tissues derived from pooled samples of two poplar plants. 5S rRNA was used as loading control.