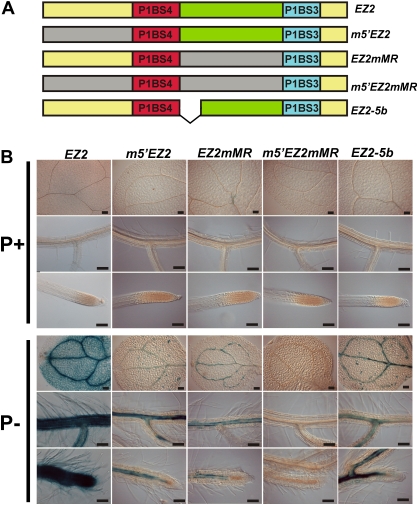
Fig. 8.
Role of the sequences adjacent to the P1BS motifs in the function of the EZ2 enhancer. (A) Diagram showing the structure of the EZ2 enhancer element and its mutant derivatives where deletions and substitutions were introduced. The region upstream of P1SB4 (m5'EZ2), the spacer sequence between P1BS4 and P1BS3 (EZ2mMR), or both the upstream and spacer sequences (m5'EZ2mMR) were replaced by random DNA sequences and the distance between the P1BS sites was reduced by 5 bp (EZ2-5b). (B) Seedlings grown for 10 days in media supplemented with 1 mM (P+) or 0 mM Pi (P–), subjected to histochemical GUS assays, and photographed using Nomarsky optics. The name of each construct is indicated at the top of each set of three images, which show the expression pattern for cotyledons, lateral roots, and root apical meristems from top to bottom, respectively. Bars, 100 μm. (This figure is available in colour at JXB online.)