Figure 4.
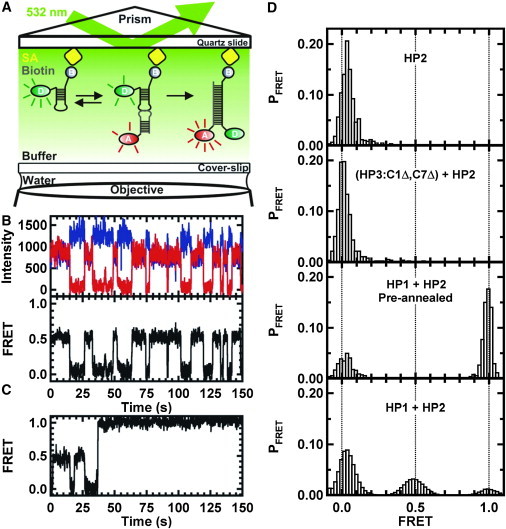
smFRET analysis of kissing kinetics. (A) Schematic representation of the total internal reflection setup. The donor RNA hairpin is immobilized by a biotin-streptavidin interaction on a biotin-BSA-coated quartz slide. A kissing interaction and an ED formation are shown in the presence of a 35-nM RNA hairpin with the acceptor fluorophore. (B) Typical single-molecule time trajectory. (Upper) Anticorrelated intensities of the donor (blue) and acceptor (red). (Lower) The corresponding FRET trajectory. Only the 0.0 and 0.5 FRET states are observed for the dissociated and kissing complexes. (C) Representative FRET trajectories showing several kissing interactions and their transition to the ED state. There are few molecules with a transition directly from KC to ED. (D) FRET histograms from single-molecule trajectories. (Upper) Donor-only RNA hairpin HP2 exhibits a single 0.0 FRET state. (Second from top) Preannealed noncomplementary hairpins (HP3-C1Δ,C7Δ + HP2) do not form the ED. (Third from top) Two preannealed complementary RNA hairpins (HP1 + HP2) show two distributions (0.0 and 1.0 FRET states) when preannealed before imaging. The distribution at 1.0 FRET state is due to ED formation. (Lower) Cumulative histogram built from >100 molecules. HP1 reacted with surface-immobilized HP2. This histogram shows three distributions at 0.0, 0.5, and 1.0 FRET states. The 0.0 FRET state is free HP2, the 0.5 FRET state represents the KC, and the 1.0 FRET state corresponds to the ED. ED molecules show no dynamic behavior and are trapped in that form under the experimental conditions.
