Figure 2.
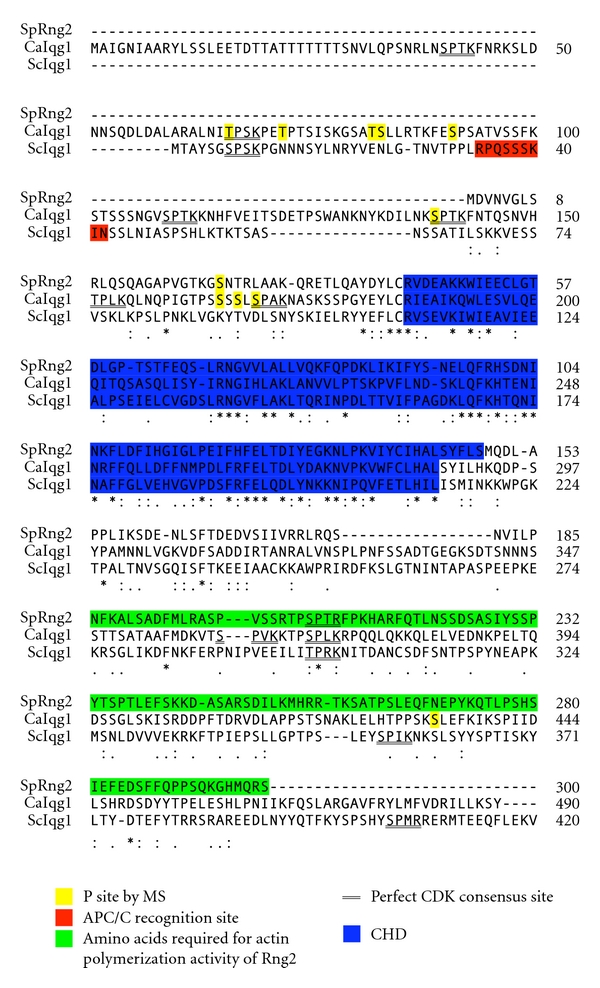
Amino acid sequence alignment of the N-terminus from S. pombe (SpRng2), C. albicans (CaIqg1) and S. cerevisiae (ScIqg1). Amino acids 1–300 of Rng2, which were identified as important for F-actin nucleation activity of Rng2 in vitro, were aligned with the N-terminal 490 amino acids of CaIqg1 and 420 amino acids of ScIqg1 using Clustal 2.1 multiple sequence alignment default settings at http://www.ebi.ac.uk/Tools/msa/clustalw2/. Amino acids highlighted in yellow were identified as phosphorylation sites by mass spec analysis, doubly underlined amino acids represent consensus CDK sites (S/TPxK), amino acids highlighted blue are the CHD, ScIqg1 amino acids highlighted in red are the APC/C recognition site, and Rng2 amino acids highlighted green are those identified as being important for F-actin nucleation activity.
