Figure 1.
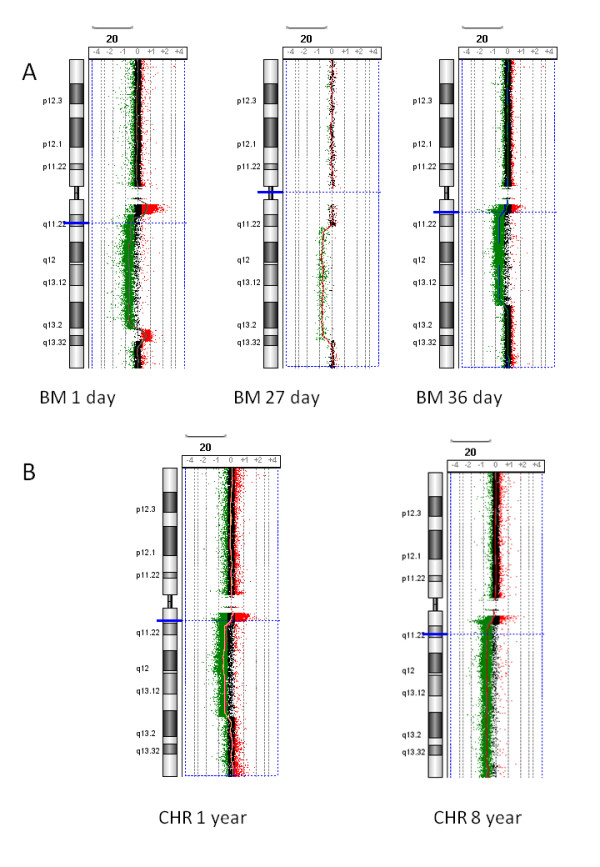
Examples of array CGH. A. Images produced using DNA extracted from bone marrow (BM) refrigerated for 1 day (diagnosis specimen of SVH05 [8]), 27 days (AML specimen from [7]) and 36 days. B. Images produced from fixed cytogenetic preparations (CHR) stored at -80°C for one year and 8 years. All of these 20q deletions were validated with the 20q12 Vysis probe, LSI D20S108 (20q12) SpectrumOrange. A 2 Mb moving average line is shown for each experiment. Each image represents duplicate experiments except for the 27 day old bone marrow, which represents one experiment. The catalog 60K array used for the BM 27 day result which has a median probe spacing of 41 kb and the other results are from a Agilent 44K custom array with probes 200 bp (20q11.21- > 20q11.22), 5 kb (20q11.22- > 20q12) and 9 kb (20p and 20q13- > 20qter) apart.
