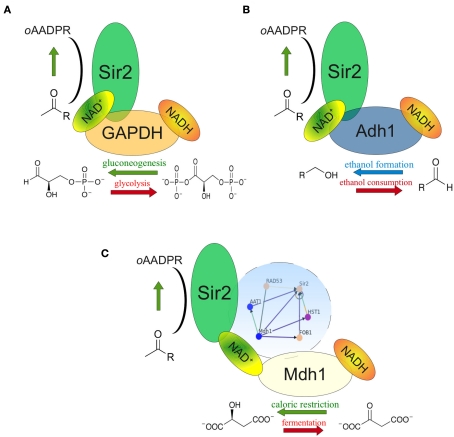
Figure 2.
NAD+ feeding or depletion in the Sir2 microenvironment. (A) The NAD(H) metabolizing glycolytic enzyme GAPDH (Tdh3) is found in the same protein complex as Sir2. At high glycolytic flux, GAPDH deprives NAD+ in close proximity to Sir2; during gluconeogenesis it generates NAD+. Genetic interplay of Tdh3 and Sir2 influences the rate of mitotic recombination. (B) The predominant cytoplasmic alcohol dehydrogenase (Adh11) also complexes with Sir2. Adh1 overexpression increased the NAD+/NADH ratio and Sir2 activity as well as prolonged yeast replicative lifespan. (C) Mitochondrial malate dehydrogenase Mdh1 is up-regulated during caloric restriction. Its overexpression increases NADH oxidation and prolongs replicative lifespan in a Sir2 dependent manner. Complex formation between Sir2 and Mdh1 has not been reported, but both proteins are interconnected through a dense genetic interaction network involving Rad53, Aat1, Fob1, and Hst1 (blue circle).