Figure 2.1.
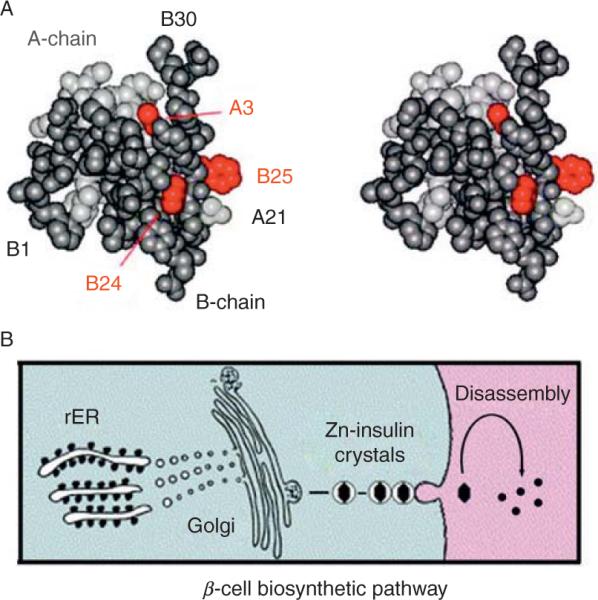
Globular structure of an insulin monomer and pathway of insulin biosyn-thesis. (A) Space-filling model of an insulin monomer highlighting sites of classical diabetes-associated mutations (red): ValA3→Leu, PheB24→ Ser, and PheB25→Leu (Shoelson et al., 1983). The A- and B-chains are otherwise shown in light and dark gray, respectively. Atomic coordinates were obtained from Protein Databank entry 4INS (2-Zn molecule 1). (B) Nascent proinsulin folds as a monomer in ER wherein zinc-ion concentration is low; in Golgi apparatus zinc-stabilized proinsulin hexamer assembles. The prohormone is processed by cleavage of connecting peptide in post-Golgi vescicles to yield mature insulin. Zinc-insulin crystals are observed in secretory granules. Insulin hexamers dissociate in the bloodstream to release active zinc-free monomers.
