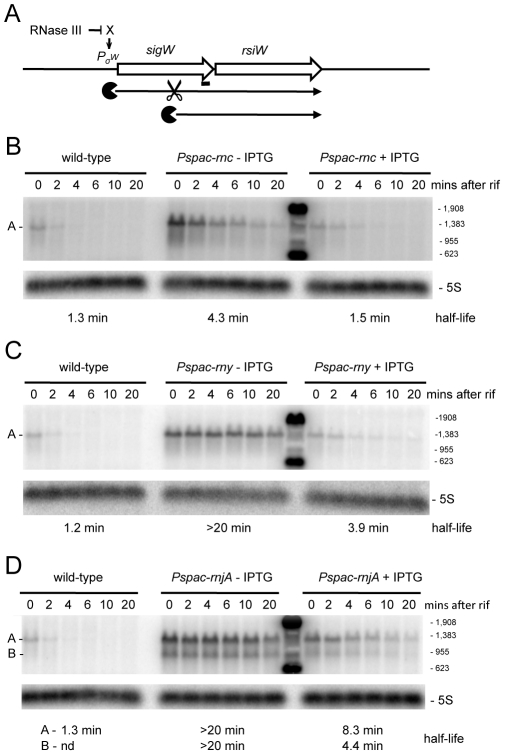
Figure 7. Degradation of the sigW-rsiW mRNA depends on RNase Y and RNase J1, while its transcription is dependent on RNase III.
(A) Structure and predicted degradation pathway of the sigW-rsiW transcript. ORFs are shown as large white arrows, transcripts as thin black arrows. Scissors indicate cleavage by RNase Y, ‘Pacman’ symbols represent 5′-3′ degradation by RNase J1. A short thick line indicates the position of the probe used. PσW indicates the approximate promoter position and relevant sigma factor mapped in [31], [32]. The schematic also depicts RNase III initiated degradation of a transcript encoding an unknown factor X early in the SigW cascade. (B) Northern blot of total mRNA isolated at times after addition of rifampicin (rif) from wild-type and RNase III depleted (Pspac-rnc−IPTG) and RNase III induced (Pspac-rnc+IPTG) cells. The blot was probed with 5′-labeled oligo CCB900 (Table S6) and reprobed with oligo HP246 against 5S rRNA. The half-life of the sigW-rsiW transcript is given under the Northern blot. Migration positions of RNA markers are shown to the right of the blot. Cross-hybridization to the marker is visible in the lane between the (−) and (+) IPTG samples. (C) Northern blot of total mRNA isolated at times after addition of rifampicin (rif) from wild-type and RNase Y depleted (Pspac-rny−IPTG) and RNase Y induced (Pspac-rny+IPTG) cells. Description as in panel (B). (D) Northern blot of total mRNA isolated at times after addition of rifampicin (rif) from wild-type and RNase J1 depleted (Pspac-rnjA−IPTG) and RNase J1 induced (Pspac-rnjA+IPTG) cells. Description as in panel (B).