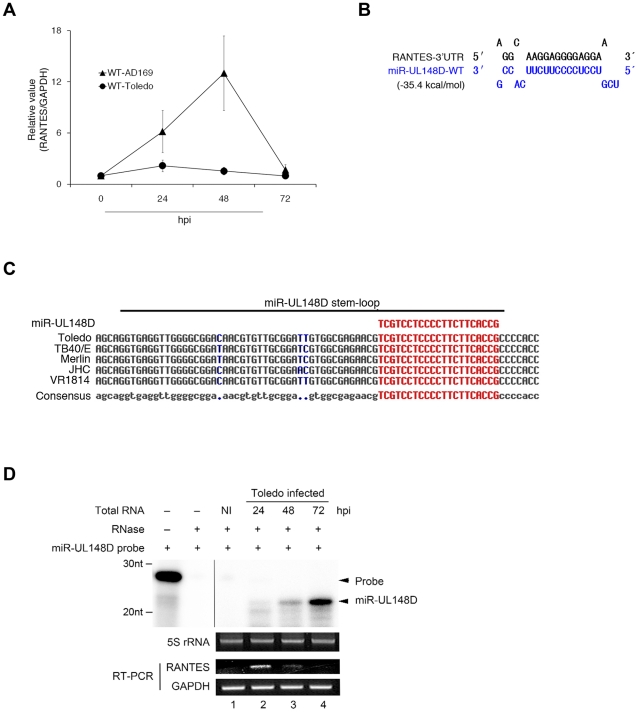
Figure 1. Inverse correlation between RANTES mRNA and miR-UL148D expression at a later stage during Toledo infection.
(A) HFF cells were infected with WT-AD169 (triangles) or WT-Toledo (circles). RANTES mRNA at indicated time of post-infection (hpi) was quantified by quantitative RT-PCR and data were normalized to GAPDH mRNA (RANTES/GAPDH). Values represent the average ± S.D. of triplicate experiments. (B) The predicted duplex of the 3′UTR of RANTES and miR-UL148D-WT. The indicated free energy value was calculated using RNA22 (http://cbcsrv.watson.ibm.com/rna22.html). (C) Alignment of miR-UL148D stem-loop sequence of various HCMV clinical strains. We analyzed the genomic sequences of several HCMV clinical strains from GenBank (Toledo GU937742.1, TB40/E AY446866.1, Merlin AY446894.2, JHC HQ380895.1, VR1814 GU179289.1). The conserved sequence (gray) and non-conserved sequence (blue) is shown. Mature miR-UL148D sequence is highlighted in red. (D) The detection of miR-UL148D in Toledo-infected HFF cells was assessed using the RNase protection assay as described under “Materials and Methods.” 5S rRNA bands stained with ethidium bromide are presented as a loading control. Using the same RNA samples, the RANTES mRNA and GAPDH mRNA were measured by RT-PCR (lower panel). NI indicates non-infected control.