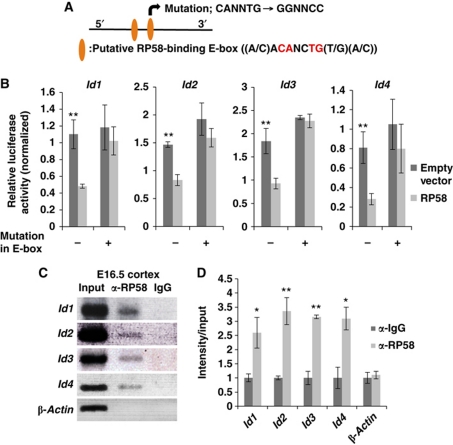
Figure 3.
RP58 directly bound to the regulatory regions of Id genes and repressed their activity. (A) Schematic shows two E-boxes (CANNTG) with specific sequences bound by RP58. The mutation was introduced into the latter E-box. Besides the typical E-boxes (CANNTG), there is only one pair of Rp58-binding E-boxes (ACANCTG) situated in close proximity to the id-encoding genomic sequences. (B) Luciferase assay representing the effects of RP58 on the putative RP58-binding region of each Id. Cos7 cells were transiently transfected with 0.2 μg pGL4P-Id1, -Id2, -Id3, or -Id4 regulatory region-luc as a reporter. The luciferase activity of cells co-transfected with Rp58 expression vector or pCDNA3.1 (+) empty vector (0.2 μg) was measured. The mean Photinus pyralis firefly activities were normalized to the mean activities of Renilla luciferase vector (0.1 μg) (t-test: **P<0.01). Data were represented as mean values±s.d. (n=3). (C) ChIP analysis of the putative RP58-binding region of each Id gene. Mouse E16.5 homogenized cerebral cortex was immunoprecipitated with an antibody specific for RP58 or rabbit-IgG as a negative control. As a positive control, 0.2% total input chromatin DNA was used, and the amplification products of the β-actin promoter region were used as a negative control. (D) The agarose gel image presented in (C) was quantified using Fiji software available online (http://pacific.mpi-cbg.de/wiki/index.php/Fiji) (t-test: *P<0.05, **P<0.01). Data were represented as mean±s.d. (n=3).